Agrobacterium tumefaciens-Mediated Genetic Transformation of the Ect-endomycorrhizal Fungus Terfezia boudieri
Abstract
:1. Introduction
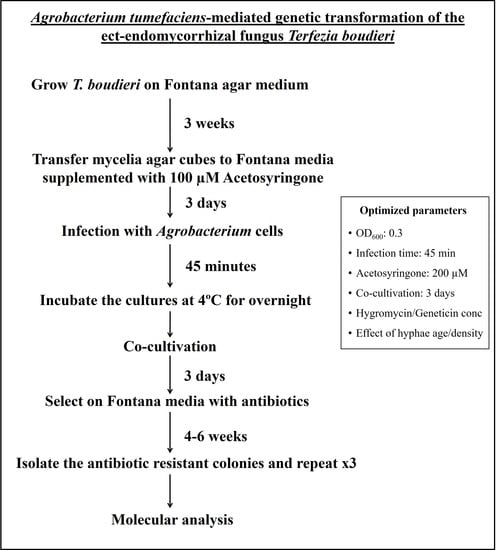
2. Materials and Methods
2.1. Terfezia Boudieri Culture
2.2. Hygromycin and Geneticin Sensitivity Test
2.3. A. tumefaciens Culture and Plasmid Transfection/Electroporation
2.4. T. boudieri Infection and Co-Cultivation with A. tumefaciens
2.5. Selection of Fungal Transformants
2.6. Microscopic Analysis
2.7. Fungal Genomic DNA Extraction and PCR Analysis
2.8. Fungal Transgene Insertion and Stability
2.9. Inoculation of the Host Plant with Transformed Hyphae
2.10. Data Collection and Statistical Analysis
3. Results
3.1. Sensitivity of T. boudieri Mycelium to Antibiotics and the Effect of Acetosyringone on Transformation Efficiency
3.2. Effects of A. tumefaciens Cell Density and Infection Time on T. boudieri Transformation
3.3. The Effect of Pre-Selection Cultivation Time on Transformation Efficiency
3.4. The Effect of the Mycelia Developmental Stage on Transformation Efficiency
3.5. Molecular Verification of Putative Transformants
3.6. Genetic Stability of T. boudieri Transformants
3.7. Expression of Reporter Genes in Free-Living Mycelia and in Mycorrhizal Association
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moreno, G.; Alvarado, P.; Manjón, J.L. Hypogeous desert fungi. In Desert Truffles; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–20. [Google Scholar]
- Kamle, M.; Bar, E.; Lewinsohn, D.; Shavit, E.; Roth-Bejerano, N.; Kagan-Zur, V.; Barak, Z.; Guy, O.; Zaady, E.; Lewinsohn, E.; et al. Characterization of Morphology, Volatile Profiles, and Molecular Markers in Edible Desert Truffles from the Negev Desert. J. Agric. Food Chem. 2017, 65, 2977–2983. [Google Scholar] [CrossRef] [PubMed]
- Zaretsky, M.; Kagan-Zur, V.; Mills, D.; Roth-Bejerano, N. Analysis of mycorrhizal associations formed by Cistus incanus transformed root clones with Terfezia boudieri isolates. Plant Cell Rep. 2005, 25, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Morte, A.; Zamora, M.; Gutierrez, A.; Honrubia, M. Desert Truffle Cultivation in Semiarid Mediterranean Areas. In Mycorrhizas-Functional Processes and Ecological Impact; Springer: Berlin/Heidelberg, Germany, 2009; pp. 221–233. [Google Scholar]
- Kovács, G.M.; Balázs, T.K.; Calonge, F.D.; Martín, M.P.; Andrade-Linares, D.R.; Grosch, R.; Franken, P.; Rexer, K.-H.; Kost, G.; Restrepo, S.; et al. The diversity of Terfezia desert truffles: New species and a highly variable species complex with intrasporocarpic nrDNA ITS heterogeneity. Mycologia 2011, 103, 841–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turgeman, T.; Ben Asher, J.; Roth-Bejerano, N.; Kagan-Zur, V.; Kapulnik, Y.; Sitrit, Y. Mycorrhizal association between the desert truffle Terfezia boudieri and Helianthemum sessiliflorum alters plant physiology and fitness to arid conditions. Mycorrhiza 2011, 21, 623–630. [Google Scholar] [CrossRef]
- Murat, C.; Payen, T.; Noel, B.; Kuo, A.; Morin, E.; Chen, J.; Kohler, A.; Krizsán, K.; Balestrini, R.; Da Silva, C.; et al. Pezizomycetes genomes reveal the molecular basis of ectomycorrhizal truffle lifestyle. Nat. Ecol. Evol. 2018, 2, 1956–1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soudzilovskaia, N.A.; Van Bodegom, P.M.; Terrer, C.; Zelfde, M.V.; McCallum, I.; McCormack, M.L.; Fisher, J.B.; Brundrett, M.C.; De Sá, N.C.; Tedersoo, L. Global mycorrhizal plant distribution linked to terrestrial carbon stocks. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kagan-Zur, V.; Zaretsky, M.; Sitrit, Y.; Roth-Bejerano, N. Hypogeous Pezizaceae: Physiology and molecular genetics. In Mycorrhiza; Springer: Berlin/Heidelberg, Germany, 2008; pp. 161–183. [Google Scholar]
- Shavit, E.; Shavit, E. The medicinal value of desert truffles. In Desert Truffles; Springer: Berlin/Heidelberg, Germany, 2014; pp. 323–340. [Google Scholar]
- Tagnamas, Z.; Bahammou, Y.; Kouhila, M.; Hilali, S.; Idlimam, A.; Lamharrar, A. Conservation of Moroccan truffle (Terfezia boudieri) using solar drying method. Renew. Energy 2020, 146, 16–24. [Google Scholar] [CrossRef]
- Al Obaydi, M.F.; Hamed, W.M.; Al Kury, L.T.; Talib, W.H. Terfezia boudieri: A Desert Truffle With Anticancer and Immunomodulatory Activities. Front. Nutr. 2020, 7, 38. [Google Scholar] [CrossRef]
- Murcia, M.A.; Martínez-Tomé, M.; Jiménez, A.M.; Vera, A.M.; Honrubia, M.; Parras, P. Antioxidant Activity of Edible Fungi (Truffles and Mushrooms): Losses during Industrial Processing. J. Food Prot. 2002, 65, 1614–1622. [Google Scholar] [CrossRef]
- Hamza, A.; Zouari, N.; Zouari, S.; Jdir, H.; Zaidi, S.; Gtari, M.; Neffati, M. Nutraceutical potential, antioxidant and antibacterial activities of Terfezia boudieri Chatin, a wild edible desert truffle from Tunisia arid zone. Arab. J. Chem. 2016, 9, 383–389. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.; Rauf, A.; Khan, H.; Khalid, S.; Mubarak, M.S. Potential health benefits of natural products derived from truffles: A review. Trends Food Sci. Technol. 2017, 70, 1–8. [Google Scholar] [CrossRef]
- Shavit, E. The history of desert truffle use. In Desert Truffles; Springer: Berlin/Heidelberg, Germany, 2014; pp. 217–241. [Google Scholar]
- Radhouani, F.; Zampieri, E.; Guasmi, F.; Ferchichi, A.; Mello, A. Assessment of β-tubulin gene as a complementary molecular marker for desert truffle identification: A study case on Tunisian samples. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2018, 153, 19–24. [Google Scholar] [CrossRef]
- Marqués-Gálvez, J.E.; Morte, A.; Navarro-Ródenas, A.; García-Carmona, F.; Pérez-Gilabert, M. Purification and characterization of Terfezia claveryi TcCAT-1, a desert truffle catalase upregulated in mycorrhizal symbiosis. PLoS ONE 2019, 14, e0219300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrino, A.; Navarro-Ródenas, A.; Marqués-Gálvez, J.E.; Morte, A. The crop of desert truffle depends on agroclimatic parameters during two key annual periods. Agron. Sustain. Dev. 2019, 39, 51. [Google Scholar] [CrossRef]
- Morte, A.; Gutiérrez, A.; Ródenas, A.N. Advances in desert truffle mycorrhization and cultivation. In Mushrooms, Humans and Nature in a Changing World; Springer: Cham, Germany, 2020; pp. 205–219. [Google Scholar]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.A.; Otillar, R.; Riley, R.; Salamov, A.A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2013, 42, D699–D704. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Cullen, D.; Goodwin, S.B.; Hibbett, D.; Jeffries, T.W.; Kubicek, C.P.; Kuske, C.; Magnuson, J.K.; Martin, F.; Spatafora, J.W.; et al. Fueling the future with fungal genomics. Mycology 2011, 2, 192–209. [Google Scholar]
- Brenna, A.; Montanini, B.; Muggiano, E.; Proietto, M.; Filetici, P.; Ottonello, S.; Ballario, P. Integrative gene transfer in the truffle Tuber borchii by Agrobacterium tumefaciens-mediated transformation. AMB Express 2014, 4, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Stone, M.; Schlagnhaufer, C.; Romaine, C.P. A Fruiting Body Tissue Method for EfficientAgrobacterium-Mediated Transformation ofAgaricus bisporus. Appl. Environ. Microbiol. 2000, 66, 4510–4513. [Google Scholar] [CrossRef] [Green Version]
- Combier, J.-P.; Melayah, D.; Raffier, C.; Gay, G.; Marmeisse, R. Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in the symbiotic ectomycorrhizal fungusHebeloma cylindrosporum. FEMS Microbiol. Lett. 2003, 220, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Medina, E.; A Robinson, K.; Bellingham-Johnstun, K.; Ianiri, G.; Laplante, C.; Fritz-Laylin, L.K.; Buchler, N.E. Genetic transformation of Spizellomyces punctatus, a resource for studying chytrid biology and evolutionary cell biology. eLife 2020, 9, e52741. [Google Scholar] [CrossRef]
- Grimaldi, B.; De Raaf, M.A.; Filetici, P.; Ottonello, S.; Ballario, P. Agrobacterium-mediated gene transfer and enhanced green fluorescent protein visualization in the mycorrhizal ascomycete Tuber borchii: A first step towards truffle genetics. Curr. Genet. 2005, 48, 69–74. [Google Scholar] [CrossRef]
- Pardo, A.G.; Hanif, M.; Raudaskoski, M.; Gorfer, M. Genetic transformation of ectomycorrhizal fungi mediated by Agrobacterium tumefaciens. Mycol. Res. 2002, 106, 132–137. [Google Scholar] [CrossRef]
- Hanif, M.; Pardo, A.G.; Gorfer, M.; Raudaskoski, M. T-DNA transfer and integration in the ectomycorrhizal fungus Suillus bovinus using hygromycin B as a selectable marker. Curr. Genet. 2002, 41, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Benjdia, M.; Avolio, M.; Voigt, B.; Menzel, D.; Pardo, A.; Frommer, W.B.; Wipf, D. Functional expression of the green fluorescent protein in the ectomycorrhizal model fungus Hebeloma cylindrosporum. Mycorrhiza 2006, 16, 437–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngari, C.; Combier, J.-P.; Doré, J.; Marmeisse, R.; Gay, G.; Melayah, D. The dominant Hc.Sdh R carboxin-resistance gene of the ectomycorrhizal fungus Hebeloma cylindrosporum as a selectable marker for transformation. Curr. Genet. 2009, 55, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Pardo, A.G.; Kemppainen, M.; Valdemoros, D.; Duplessis, S.; Martin, F.; Tagu, D. T-DNA transfer from Agrobacterium tumefaciens to the ectomycorrhizal fungus Pisolithus microcarpus. Rev. Argent. Microbiol. 2005, 37, 69–72. [Google Scholar]
- Kemppainen, M.; Circosta, A.; Tagu, D.; Martin, F.; Pardo, A.G. Agrobacterium-mediated transformation of the ectomycorrhizal symbiont Laccaria bicolor S238N. Mycorrhiza 2005, 16, 19–22. [Google Scholar] [CrossRef]
- Kemppainen, M.; Duplessis, S.; Martin, F.; Pardo, A.G. T-DNA insertion, plasmid rescue and integration analysis in the model mycorrhizal fungus Laccaria bicolor. Microb. Biotechnol. 2008, 1, 258–269. [Google Scholar] [CrossRef] [Green Version]
- Kemppainen, M.; Pardo, A.G. Transformation of the mycorrhizal fungus Laccaria bicolor using Agrobacterium tumefaciens. Bioeng. Bugs 2011, 2, 38–44. [Google Scholar] [CrossRef]
- Xu, H.; Kemppainen, M.; El Kayal, W.; Lee, S.H.; Pardo, A.G.; Cooke, J.E.K.; Zwiazek, J. Overexpression ofLaccaria bicoloraquaporinJQ585595alters root water transport properties in ectomycorrhizal white spruce (Picea glauca) seedlings. New Phytol. 2014, 205, 757–770. [Google Scholar] [CrossRef]
- Stephan, B.I.; Crespo, M.C.A.; Kemppainen, M.J.; Pardo, A.G. Agrobacterium-mediated insertional mutagenesis in the mycorrhizal fungus Laccaria bicolor. Curr. Genet. 2016, 63, 215–227. [Google Scholar] [CrossRef]
- Zubieta, M.P.; Coelho, I.D.S.; De Queiroz, M.V.; De Araújo, E.F. Agrobacterium tumefaciens-mediated genetic transformation of the ectomycorrhizal fungus Laccaria laccata. Ann. Microbiol. 2014, 64, 1875–1878. [Google Scholar] [CrossRef]
- Marmeisse, R.; Gay, G.; Debaud, J.-C.; Casselton, L.A. Genetic transformation of the symbiotic basidiomycete fungus Hebeloma cylindrosporum. Curr. Genet. 1992, 22, 41–45. [Google Scholar] [CrossRef]
- Forbes, P.; Millam, S.; Hooker, J.; Harrier, L. Transformation of the arbuscular mycorrhiza Gigaspora rosea by particle bombardment. Mycol. Res. 1998, 102, 497–501. [Google Scholar] [CrossRef]
- Rodriguez-Tovar, A.V.; Ruiz-Medrano, R.; Herrera-Martinez, A.; Barrera-Figueroa, B.E.; Hidalgo-Lara, M.E.; Reyes-Márquez, B.E.; Cabrera-Ponce, J.L.; Valdés, M.; Xoconostle-Cazares, B. Stable genetic transformation of the ectomycorrhizal fungus Pisolithus tinctorius. J. Microbiol. Methods 2005, 63, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.D.F.; Veneault-Fourrey, C.; Vion, P.; Guinet, F.; Morin, E.; Barry, K.W.; Lipzen, A.; Singan, V.; Pfister, S.; Na, H.; et al. Secretome Analysis from the Ectomycorrhizal Ascomycete Cenococcum geophilum. Front. Microbiol. 2018, 9, 141. [Google Scholar] [CrossRef] [Green Version]
- Bonfante, P.F. Sulla nutrizione del micelio di Tuber melanosporum Vitt in coltura. Atti Accad. Sci. Torino 1973, 107, 713–741. [Google Scholar]
- Weigel, D.; Glazebrook, J. Transformation of Agrobacterium Using Electroporation. Cold Spring Harb. Protoc. 2006, 2006, 113. [Google Scholar] [CrossRef]
- Gong, X.; Hurtado, O.; Wang, B.; Wu, C.; Yi, M.; Giraldo, M.; Valent, B.; Goodin, M.M.; Farman, M. pFPL Vectors for High-Throughput Protein Localization in Fungi: Detecting Cytoplasmic Accumulation of Putative Effector Proteins. Mol. Plant-Microbe Interact. 2015, 28, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Balaji, B.; Poulin, M.J.; Vierheilig, H.; Piché, Y. Responses of an Arbuscular Mycorrhizal Fungus, Gigaspora margarita, to Exudates and Volatiles from the Ri T-DNA-Transformed Roots of Nonmycorrhizal and Mycorrhizal Mutants of Pisum sativum L Sparkle. Exp. Mycol. 1995, 19, 275–283. [Google Scholar] [CrossRef]
- Mishra, N.C.; Tatum, E.L. Non-Mendelian Inheritance of DNA-Induced Inositol Independence in Neurospora. Proc. Natl. Acad. Sci. USA 1973, 70, 3875–3879. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.-Q.; Liu, G.-N.; Ji, R.-Y.; Shi, K.; Song, P.; Ren, L.; Huang, H.; Ji, X.-J. CRISPR/Cas9-based genome editing of the filamentous fungi: The state of the art. Appl. Microbiol. Biotechnol. 2017, 101, 7435–7443. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Tang, Y.; Lin, J.; Cai, W. Methods for genetic transformation of filamentous fungi. Microb. Cell Factories 2017, 16, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, E.R.; Hankeln, T. Transgenic Organisms and Biosafety: Horizontal Gene Transfer, Stability of DNA, and Expression of Transgenes; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Ruiz-Diez, B. Strategies for the transformation of filamentous fungi. J. Appl. Microbiol. 2002, 92, 189–195. [Google Scholar] [CrossRef]
- Weyda, I.; Yang, L.; Vang, J.; Ahring, B.K.; Lübeck, M.; Lübeck, P.S. A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J. Microbiol. Methods 2017, 135, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Weld, R.J.; Plummer, K.M.; Carpenter, M.A.; Ridgway, H.J. Approaches to functional genomics in filamentous fungi. Cell Res. 2006, 16, 31–44. [Google Scholar] [CrossRef] [Green Version]
- Utami, E.S.W.; Hariyanto, S.; Manuhara, Y.S.W. Agrobacterium tumefaciens-mediated transformation of Dendrobium lasianthera JJ Sm: An important medicinal orchid. J. Genet. Eng. Biotechnol. 2018, 16, 703–709. [Google Scholar] [CrossRef]
- Turgeman, T.; Lubinsky, O.; Roth-Bejerano, N.; Kagan-Zur, V.; Kapulnik, Y.; Koltai, H.; Zaady, E.; Ben-Shabat, S.; Guy, O.; Lewinsohn, E.; et al. The role of pre-symbiotic auxin signaling in ectendomycorrhiza formation between the desert truffle Terfezia boudieri and Helianthemum sessiliflorum. Mycorrhiza 2015, 26, 287–297. [Google Scholar] [CrossRef]
- Mello, A.; Balestrini, R. Recent Insights on Biological and Ecological Aspects of Ectomycorrhizal Fungi and Their Interactions. Front. Microbiol. 2018, 9, 216. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satish, L.; Kamle, M.; Keren, G.; Patil, C.D.; Yehezkel, G.; Barak, Z.; Kagan-Zur, V.; Kushmaro, A.; Sitrit, Y. Agrobacterium tumefaciens-Mediated Genetic Transformation of the Ect-endomycorrhizal Fungus Terfezia boudieri. Genes 2020, 11, 1293. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11111293
Satish L, Kamle M, Keren G, Patil CD, Yehezkel G, Barak Z, Kagan-Zur V, Kushmaro A, Sitrit Y. Agrobacterium tumefaciens-Mediated Genetic Transformation of the Ect-endomycorrhizal Fungus Terfezia boudieri. Genes. 2020; 11(11):1293. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11111293
Chicago/Turabian StyleSatish, Lakkakula, Madhu Kamle, Guy Keren, Chandrashekhar D. Patil, Galit Yehezkel, Ze’ev Barak, Varda Kagan-Zur, Ariel Kushmaro, and Yaron Sitrit. 2020. "Agrobacterium tumefaciens-Mediated Genetic Transformation of the Ect-endomycorrhizal Fungus Terfezia boudieri" Genes 11, no. 11: 1293. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11111293