Genomic and Transcriptomic Analysis for Identification of Genes and Interlinked Pathways Mediating Artemisinin Resistance in Leishmania donovani
Abstract
:1. Introduction
2. Materials and Methods
2.1. Parasite and Culture Condition
2.2. Genomic DNA Isolation from Parasite Culture
2.3. Genomic Library Preparation and Sequencing
2.4. Whole-Genome Sequencing Data Analysis
2.5. Functional Annotation and Classification of Unigenes
2.6. Total RNA Isolation from Parasites
2.7. Oligonucleotide Array
2.8. RNA Labelling, Amplification, Hybridization, and Data Analysis
2.9. Data Availability
2.10. Quantitative Real-Time PCR (qPCR)
2.11. Western Blotting of Promastigote Cell Lysate
2.12. Cytotoxicity Assay
2.13. Artesunate Susceptibility in the Presence of Inhibitors
2.14. Ethics Approval
3. Results
3.1. Whole-Genome Sequence Diversity Data of K133AS-R Compared to K133WT
3.2. Differentially Expressed Genes in K133AS-R vs. K133WT
3.3. Chromosomal Diversity in Artesunate-Resistant L. donovani
3.3.1. Chromosome Copy Number Variation (CNV)
3.3.2. Variance in Allelic Frequency Due to Change in Chromosomal Somy/Ploidy
3.4. Functional Annotation and Classification of K133WT and K133AS-R Unigenes
3.5. Comparative Transcriptome Analysis of K133WT vs. K133AS-R Parasites
3.6. Validation of Modulated Gene Expression Using qPCR
3.7. Targeted Protein Profiling of AQP1 and HSP70 in K133WT and K133AS-R Leishmania Parasites by Western Blotting
3.8. Susceptibility of K133WT and K133AS-R Parasites in the Presence of the AQP1 Inhibitor and Modulator of ABC Transporters
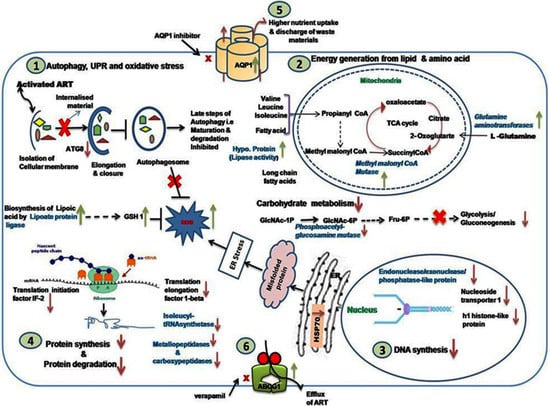
3.9. Analysis of Modulated Genes and Pathways in K133AS-R Parasites
3.9.1. Autophagy, UPR, and Oxidative Stress
3.9.2. Carbohydrate, Lipid, and Amino Acid Metabolism
3.9.3. DNA Synthesis and Translation Machinery
3.9.4. Modulated Expression of Transporters
3.10. Correlation of Whole-Genome Sequencing Analysis with Transcriptomic Data
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis [seminar]. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Alvar, J.; Vélez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; de Boer, M. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef] [PubMed]
- Torres-Guerrero, E.; Quintanilla-Cedillo, M.R.; Ruiz-Esmenjaud, J.; Arenas, R. Leishmaniasis: A review. F1000Research 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Salih, N.A.; van Griensven, J.; Chappuis, F.; Antierens, A.; Mumina, A.; Hammam, O.; Boulle, P.; Alirol, E.; Alnour, M.; Elhag, M.S. Liposomal amphotericin B for complicated visceral leishmaniasis (kala-azar) in eastern Sudan: How effective is treatment for this neglected disease? Trop. Med. Int. Health 2014, 19, 146–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rijal, S.; Ostyn, B.; Uranw, S.; Rai, K.; Bhattarai, N.R.; Dorlo, T.P.C.; Beijnen, J.H.; Vanaerschot, M.; Decuypere, S.; Dhakal, S.S. Increasing failure of miltefosine in the treatment of Kala-azar in Nepal and the potential role of parasite drug resistance, reinfection, or noncompliance. Clin. Infect. Dis. 2013, 56, 1530–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundar, S.; Singh, A.; Rai, M.; Prajapati, V.K.; Singh, A.K.; Ostyn, B.; Boelaert, M.; Dujardin, J.-C.; Chakravarty, J. Efficacy of miltefosine in the treatment of visceral leishmaniasis in India after a decade of use. Clin. Infect. Dis. 2012, 55, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Islamuddin, M.; Chouhan, G.; Want, M.Y.; Tyagi, M.; Abdin, M.Z.; Sahal, D.; Afrin, F. Leishmanicidal activities of Artemisia annua leaf essential oil against visceral leishmaniasis. Front. Microbiol. 2014, 5, 626. [Google Scholar] [CrossRef] [Green Version]
- Ghaffarifar, F.; Heydari, F.E.; Dalimi, A.; Hassan, Z.M.; Delavari, M.; Mikaeiloo, H. Evaluation of apoptotic and antileishmanial activities of Artemisinin on promastigotes and BALB/C mice infected with Leishmania major. Iran. J. Parasitol. 2015, 10, 258. [Google Scholar]
- Want, M.Y.; Islamuddin, M.; Chouhan, G.; Ozbak, H.A.; Hemeg, H.A.; Dasgupta, A.K.; Chattopadhyay, A.P.; Afrin, F. Therapeutic efficacy of artemisinin-loaded nanoparticles in experimental visceral leishmaniasis. Colloids Surf. B Biointerfaces 2015, 130, 215–221. [Google Scholar] [CrossRef]
- Sen, R.; Bandyopadhyay, S.; Dutta, A.; Mandal, G.; Ganguly, S.; Saha, P.; Chatterjee, M. Artemisinin triggers induction of cell-cycle arrest and apoptosis in Leishmania donovani promastigotes. J. Med. Microbiol. 2007, 56, 1213–1218. [Google Scholar] [CrossRef] [Green Version]
- Ebrahimisadr, P.; Ghaffarifar, F.; Hassan, Z.M. In-vitro evaluation of antileishmanial activity and toxicity of artemether with focus on its apoptotic effect. Iran. J. Pharm. Res. IJPR 2013, 12, 903. [Google Scholar] [PubMed]
- Nemati, S.; Nahrevanian, H.; Haniloo, A.; Farahmand, M. Investigation on nitric oxide and C-reactive protein involvement in antileishmanial effects of artemisinin and glucantim on cutaneous leishmaniasis. Adv. Stud. Biol. 2013, 5, 27–36. [Google Scholar] [CrossRef]
- Rodrigues, I.A.; Mazotto, A.M.; Cardoso, V.; Alves, R.L.; Amaral, A.C.F.; Silva, J.R.D.A.; Pinheiro, A.S.; Vermelho, A.B. Natural products: Insights into leishmaniasis inflammatory response. Mediat. Inflamm. 2015, 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, R.; Saha, P.; Sarkar, A.; Ganguly, S.; Chatterjee, M. Iron enhances generation of free radicals by Artemisinin causing a caspase-independent, apoptotic death in Leishmania donovani promastigotes. Free Radic. Res. 2010, 44, 1289–1295. [Google Scholar] [CrossRef]
- Hefnawy, A.; Berg, M.; Dujardin, J.-C.; De Muylder, G. Exploiting knowledge on Leishmania drug resistance to support the quest for new drugs. Trends Parasitol. 2017, 33, 162–174. [Google Scholar] [CrossRef] [Green Version]
- Ouellette, M.; Drummelsmith, J.; Papadopoulou, B. Leishmaniasis: Drugs in the clinic, resistance and new developments. Drug Resist. Updat. 2004, 7, 257–266. [Google Scholar] [CrossRef]
- Pourshafie, M.; Morand, S.; Virion, A.; Rakotomanga, M.; Dupuy, C.; Loiseau, P.M. Cloning of S-adenosyl-l-methionine: C-24-Δ-sterol-methyltransferase (ERG6) from Leishmania donovani and characterization of mRNAs in wild-type and amphotericin B-resistant promastigotes. Antimicrob. Agents Chemother. 2004, 48, 2409–2414. [Google Scholar] [CrossRef] [Green Version]
- Jhingran, A.; Chawla, B.; Saxena, S.; Barrett, M.P.; Madhubala, R. Paromomycin: Uptake and resistance in Leishmania donovani. Mol. Biochem. Parasitol. 2009, 164, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Castanys-Muñoz, E.; Alder-Baerens, N.; Pomorski, T.; Gamarro, F.; Castanys, S. A novel ATP-binding cassette transporter from Leishmania is involved in transport of phosphatidylcholine analogues and resistance to alkyl-phospholipids. Mol. Microbiol. 2007, 64, 1141–1153. [Google Scholar] [CrossRef]
- Mandal, S.; Maharjan, M.; Singh, S.; Chatterjee, M.; Madhubala, R. Assessing aquaglyceroporin gene status and expression profile in antimony-susceptible and-resistant clinical isolates of Leishmania donovani from India. J. Antimicrob. Chemother. 2010, 65, 496–507. [Google Scholar] [CrossRef] [Green Version]
- Maltezou, H.C. Drug resistance in visceral leishmaniasis. J. Biomed. Biotechnol. 2009, 2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deep, D.K.; Singh, R.; Bhandari, V.; Verma, A.; Sharma, V.; Wajid, S.; Sundar, S.; Ramesh, V.; Dujardin, J.C.; Salotra, P. Increased miltefosine tolerance in clinical isolates of Leishmania donovani is associated with reduced drug accumulation, increased infectivity and resistance to oxidative stress. PLoS Negl. Trop. Dis. 2017, 11, e0005641. [Google Scholar] [CrossRef] [PubMed]
- Kulshrestha, A.; Sharma, V.; Singh, R.; Salotra, P. Comparative transcript expression analysis of miltefosine-sensitive and miltefosine-resistant Leishmania donovani. Parasitol. Res. 2014, 113, 1171–1184. [Google Scholar] [CrossRef]
- Verma, A.; Bhandari, V.; Deep, D.K.; Sundar, S.; Dujardin, J.C.; Singh, R.; Salotra, P. Transcriptome profiling identifies genes/pathways associated with experimental resistance to paromomycin in Leishmania donovani. Int. J. Parasitol. Drugs Drug Resist. 2017, 7, 370–377. [Google Scholar] [CrossRef]
- Ubeda, J.-M.; Légaré, D.; Raymond, F.; Ouameur, A.A.; Boisvert, S.; Rigault, P.; Corbeil, J.; Tremblay, M.J.; Olivier, M.; Papadopoulou, B. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008, 9, R115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leprohon, P.; Legare, D.; Raymond, F.; Madore, E.; Hardiman, G.; Corbeil, J.; Ouellette, M. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 2009, 37, 1387–1399. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.; Langston, L.D.; Ouellette, M. Intrachromosomal tandem duplication and repeat expansion during attempts to inactivate the subtelomeric essential gene GSH1 in Leishmania. Nucleic Acids Res. 2011, 39, 7499–7511. [Google Scholar] [CrossRef] [Green Version]
- Ouameur, A.A.; Girard, I.; Légaré, D.; Ouellette, M. Functional analysis and complex gene rearrangements of the folate/biopterin transporter (FBT) gene family in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 2008, 162, 155–164. [Google Scholar] [CrossRef]
- Downing, T.; Imamura, H.; Decuypere, S.; Clark, T.G.; Coombs, G.H.; Cotton, J.A.; Hilley, J.D.; de Doncker, S.; Maes, I.; Mottram, J.C. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011, 21, 2143–2156. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.; Boisvert, S.; do Monte-Neto, R.L.; Coelho, A.C.; Raymond, F.; Mukhopadhyay, R.; Corbeil, J.; Ouellette, M. Telomeric gene deletion and intrachromosomal amplification in antimony-resistant L eishmania. Mol. Microbiol. 2013, 88, 189–202. [Google Scholar] [CrossRef]
- Kumar, P.; Lodge, R.; Raymond, F.; Ritt, J.; Jalaguier, P.; Corbeil, J.; Ouellette, M.; Tremblay, M.J. Gene expression modulation and the molecular mechanisms involved in Nelfinavir resistance in Leishmania donovani axenic amastigotes. Mol. Microbiol. 2013, 89, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Leprohon, P.; Fernandez-Prada, C.; Gazanion, É.; Monte-Neto, R.; Ouellette, M. Drug resistance analysis by next generation sequencing in Leishmania. Int. J. Parasitol. Drugs Drug Resist. 2015, 5, 26–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Victoria, F.J.; Castanys, S.; Gamarro, F. Leishmania donovani resistance to miltefosine involves a defective inward translocation of the drug. Antimicrob. Agents Chemother. 2003, 47, 2397–2403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Victoria, F.J.; Sánchez-Cañete, M.P.; Castanys, S.; Gamarro, F. Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J. Biol. Chem. 2006, 281, 23766–23775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, K.; Pérez-Victoria, F.J.; Stettler, M.; Sánchez-Cañete, M.P.; Castanys, S.; Gamarro, F.; Croft, S.L. Inactivation of the miltefosine transporter, LdMT, causes miltefosine resistance that is conferred to the amastigote stage of Leishmania donovani and persists in vivo. Int. J. Antimicrob. Agents 2007, 30, 229–235. [Google Scholar] [CrossRef]
- Srivastava, S.; Mishra, J.; Gupta, A.K.; Singh, A.; Shankar, P.; Singh, S. Laboratory confirmed miltefosine resistant cases of visceral leishmaniasis from India. Parasites Vectors 2017, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Cojean, S.; Houzé, S.; Haouchine, D.; Huteau, F.; Lariven, S.; Hubert, V.; Michard, F.; Bories, C.; Pratlong, F.; Le Bras, J. Leishmania resistance to miltefosine associated with genetic marker. Emerg. Infect. Dis. 2012, 18, 704. [Google Scholar] [CrossRef]
- Mondelaers, A.; Sanchez-Cañete, M.P.; Hendrickx, S.; Eberhardt, E.; Garcia-Hernandez, R.; Lachaud, L.; Cotton, J.; Sanders, M.; Cuypers, B.; Imamura, H. Genomic and molecular characterization of miltefosine resistance in Leishmania infantum strains with either natural or acquired resistance through experimental selection of intracellular amastigotes. PLoS ONE 2016, 11, e0154101. [Google Scholar] [CrossRef]
- Takala-Harrison, S.; Clark, T.G.; Jacob, C.G.; Cummings, M.P.; Miotto, O.; Dondorp, A.M.; Fukuda, M.M.; Nosten, F.; Noedl, H.; Imwong, M. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc. Natl. Acad. Sci. USA 2013, 110, 240–245. [Google Scholar] [CrossRef] [Green Version]
- Ariey, F.; Witkowski, B.; Amaratunga, C.; Beghain, J.; Langlois, A.-C.; Khim, N.; Kim, S.; Duru, V.; Bouchier, C.; Ma, L. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014, 505, 50–55. [Google Scholar] [CrossRef]
- Verma, A.; Ghosh, S.; Salotra, P.; Singh, R. Artemisinin-resistant Leishmania parasite modulates host cell defense mechanism and exhibits altered expression of unfolded protein response genes. Parasitol. Res. 2019, 118, 2705–2713. [Google Scholar] [CrossRef] [PubMed]
- Bronner, I.F.; Quail, M.A.; Turner, D.J.; Swerdlow, H. Improved protocols for illumina sequencing. Curr. Protoc. Hum. Genet. 2013, 79, 12–18. [Google Scholar] [CrossRef]
- Dumetz, F.; Imamura, H.; Sanders, M.; Seblova, V.; Myskova, J.; Pescher, P.; Vanaerschot, M.; Meehan, C.J.; Cuypers, B.; De Muylder, G. Modulation of aneuploidy in Leishmania donovani during adaptation to different in vitro and in vivo environments and its impact on gene expression. MBio 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wysoker, A. Durbin R; 1000 Genome project data processing subgroup. The sequence alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Kent, W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, A.; Yamazaki, M.; Takahashi, H.; Nakamura, M.; Kojoma, M.; Suzuki, H.; Saito, K. RNA-seq transcriptome analysis of Panax japonicus, and its comparison with other Panax species to identify potential genes involved in the saponins biosynthesis. Front. Plant Sci. 2016, 7, 481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consortium, G.O. Creating the gene ontology resource: Design and implementation. Genome Res. 2001, 11, 1425–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Rochette, A.; McNicoll, F.; Girard, J.; Breton, M.; Leblanc, É.; Bergeron, M.G.; Papadopoulou, B. Characterization and developmental gene regulation of a large gene family encoding amastin surface proteins in Leishmania spp. Mol. Biochem. Parasitol. 2005, 140, 205–220. [Google Scholar] [CrossRef]
- Leprohon, P.; Légaré, D.; Ouellette, M. Intracellular localization of the ABCC proteins of Leishmania and their role in resistance to antimonials. Antimicrob. Agents Chemother. 2009, 53, 2646–2649. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, V.; Sundar, S.; Dujardin, J.C.; Salotra, P. Elucidation of cellular mechanisms involved in experimental paromomycin resistance in Leishmania donovani. Antimicrob. Agents Chemother. 2014, 58, 2580–2585. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.; Sharma, P.; Selvapandiyan, A.; Salotra, P. Leishmania donovani-specific U b-related modifier-1: An early endosome-associated ubiquitin-like conjugation in Leishmania donovani. Mol. Microbiol. 2016, 99, 597–610. [Google Scholar] [CrossRef]
- Samarasinghe, S.R.; Samaranayake, N.; Kariyawasam, U.L.; Siriwardana, Y.D.; Imamura, H.; Karunaweera, N.D. Genomic insights into virulence mechanisms of Leishmania donovani: Evidence from an atypical strain. BMC Genom. 2018, 19, 843. [Google Scholar] [CrossRef]
- Rakotomanga, M.; Saint-Pierre-Chazalet, M.; Loiseau, P.M. Alteration of fatty acid and sterol metabolism in miltefosine-resistant Leishmania donovani promastigotes and consequences for drug-membrane interactions. Antimicrob. Agents Chemother. 2005, 49, 2677–2686. [Google Scholar] [CrossRef] [Green Version]
- Besteiro, S.; Tonn, D.; Tetley, L.; Coombs, G.H.; Mottram, J.C. The AP3 adaptor is involved in the transport of membrane proteins to acidocalcisomes of Leishmania. J. Cell Sci. 2008, 121, 561–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tonkin, M.L.; Roques, M.; Lamarque, M.H.; Pugnière, M.; Douguet, D.; Crawford, J.; Lebrun, M.; Boulanger, M.J. Host cell invasion by apicomplexan parasites: Insights from the co-structure of AMA1 with a RON2 peptide. Science 2011, 333, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Camacho, E.; González-De la Fuente, S.; Rastrojo, A.; Peiró-Pastor, R.; Solana, J.C.; Tabera, L.; Gamarro, F.; Carrasco-Ramiro, F.; Requena, J.M.; Aguado, B. Complete assembly of the Leishmania donovani (HU3 strain) genome and transcriptome annotation. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Lustigman, S.; Zhang, J.; Liu, J.; Oksov, Y.; Hashmi, S. RNA interference targeting cathepsin L and Z-like cysteine proteases of Onchocerca volvulus confirmed their essential function during L3 molting. Mol. Biochem. Parasitol. 2004, 138, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Dalton, J.P.; Neill, S.O.; Stack, C.; Collins, P.; Walshe, A.; Sekiya, M.; Doyle, S.; Mulcahy, G.; Hoyle, D.; Khaznadji, E. Fasciola hepatica cathepsin L-like proteases: Biology, function, and potential in the development of first generation liver fluke vaccines. Int. J. Parasitol. 2003, 33, 1173–1181. [Google Scholar] [CrossRef]
- Appalasamy, S.; Lo, K.Y.; Ch’ng, S.J.; Nornadia, K.; Othman, A.S.; Chan, L.-K. Antimicrobial activity of artemisinin and precursor derived from in vitro plantlets of Artemisia annua L. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; He, R.; Kapoor, A.; Forman, M.; Mazzone, J.R.; Posner, G.H.; Arav-Boger, R. Inhibition of human cytomegalovirus replication by artemisinins: Effects mediated through cell cycle modulation. Antimicrob. Agents Chemother. 2015, 59, 3870–3879. [Google Scholar] [CrossRef] [Green Version]
- Want, M.Y.; Islammudin, M.; Chouhan, G.; Ozbak, H.A.; Hemeg, H.A.; Chattopadhyay, A.P.; Afrin, F. Nanoliposomal artemisinin for the treatment of murine visceral leishmaniasis. Int. J. Nanomed. 2017, 12, 2189. [Google Scholar] [CrossRef] [Green Version]
- Spielmann, M.; Kakar, N.; Tayebi, N.; Leettola, C.; Nürnberg, G.; Sowada, N.; Lupiáñez, D.G.; Harabula, I.; Flöttmann, R.; Horn, D. Exome sequencing and CRISPR/Cas genome editing identify mutations of ZAK as a cause of limb defects in humans and mice. Genome Res. 2016, 26, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Günzl, A. RNA polymerases and transcription factors of trypanosomes. In RNA Metabolism in Trypanosomes; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–27. [Google Scholar]
- Barja, P.P.; Pescher, P.; Bussotti, G.; Dumetz, F.; Imamura, H.; Kedra, D.; Domagalska, M.; Chaumeau, V.; Himmelbauer, H.; Pages, M. Haplotype selection as an adaptive mechanism in the protozoan pathogen Leishmania donovani. Nat. Ecol. Evol. 2017, 1, 1961–1969. [Google Scholar] [CrossRef]
- Gilissen, C.; Hehir-Kwa, J.Y.; Thung, D.T.; van de Vorst, M.; van Bon, B.W.M.; Willemsen, M.H.; Kwint, M.; Janssen, I.M.; Hoischen, A.; Schenck, A. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014, 511, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kedzierski, L.; Montgomery, J.; Bullen, D.; Curtis, J.; Gardiner, E.; Jimenez-Ruiz, A.; Handman, E. A leucine-rich repeat motif of Leishmania parasite surface antigen 2 binds to macrophages through the complement receptor 3. J. Immunol. 2004, 172, 4902–4906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gokulasuriyan, R.K.; Ghosh, M. Comparative in-silico genome analysis of Leishmania (Leishmania) donovani: A step towards its species specificity. Meta Gene 2014, 2, 782–798. [Google Scholar]
- Laha, B.; Verma, A.K.; Biswas, B.; Sengodan, S.K.; Rastogi, A.; Willard, B.; Ghosh, M. Detection and characterization of an albumin-like protein in Leishmania donovani. Parasitol. Res. 2019, 118, 1609–1623. [Google Scholar] [CrossRef]
- Jackson, A.P. The evolution of amastin surface glycoproteins in trypanosomatid parasites. Mol. Biol. Evol. 2010, 27, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Bridgford, J.L.; Xie, S.C.; Cobbold, S.A.; Pasaje, C.F.A.; Herrmann, S.; Yang, T.; Gillett, D.L.; Dick, L.R.; Ralph, S.A.; Dogovski, C. Artemisinin kills malaria parasites by damaging proteins and inhibiting the proteasome. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Beitz, E. Aquaporins from pathogenic protozoan parasites: Structure, function and potential for chemotherapy. Biol. Cell 2005, 97, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Marquis, N.; Gourbal, B.; Rosen, B.P.; Mukhopadhyay, R.; Ouellette, M. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol. Microbiol. 2005, 57, 1690–1699. [Google Scholar] [CrossRef]
- Kumar, D.; Singh, R.; Bhandari, V.; Kulshrestha, A.; Negi, N.S.; Salotra, P. Biomarkers of antimony resistance: Need for expression analysis of multiple genes to distinguish resistance phenotype in clinical isolates of Leishmania donovani. Parasitol. Res. 2012, 111, 223–230. [Google Scholar] [CrossRef]
- Leprohon, P.; Légaré, D.; Girard, I.; Papadopoulou, B.; Ouellette, M. Modulation of Leishmania ABC protein gene expression through life stages and among drug-resistant parasites. Eukaryot. Cell 2006, 5, 1713–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purkait, B.; Kumar, A.; Nandi, N.; Sardar, A.H.; Das, S.; Kumar, S.; Pandey, K.; Ravidas, V.; Kumar, M.; De, T. Mechanism of amphotericin B resistance in clinical isolates of Leishmania donovani. Antimicrob. Agents Chemother. 2012, 56, 1031–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]












Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghosh, S.; Verma, A.; Kumar, V.; Pradhan, D.; Selvapandiyan, A.; Salotra, P.; Singh, R. Genomic and Transcriptomic Analysis for Identification of Genes and Interlinked Pathways Mediating Artemisinin Resistance in Leishmania donovani. Genes 2020, 11, 1362. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11111362
Ghosh S, Verma A, Kumar V, Pradhan D, Selvapandiyan A, Salotra P, Singh R. Genomic and Transcriptomic Analysis for Identification of Genes and Interlinked Pathways Mediating Artemisinin Resistance in Leishmania donovani. Genes. 2020; 11(11):1362. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11111362
Chicago/Turabian StyleGhosh, Sushmita, Aditya Verma, Vinay Kumar, Dibyabhaba Pradhan, Angamuthu Selvapandiyan, Poonam Salotra, and Ruchi Singh. 2020. "Genomic and Transcriptomic Analysis for Identification of Genes and Interlinked Pathways Mediating Artemisinin Resistance in Leishmania donovani" Genes 11, no. 11: 1362. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11111362