Current and Emerging Methods for the Synthesis of Single-Stranded DNA
Abstract
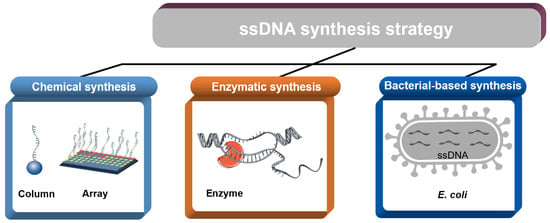
:1. Introduction
2. Chemical Synthesis
2.1. Column-Based Oligo Synthesis
2.2. Array-Based Oligo Synthesis
3. Enzymatic Synthesis
3.1. Terminal Deoxynucleotidyl Transferase
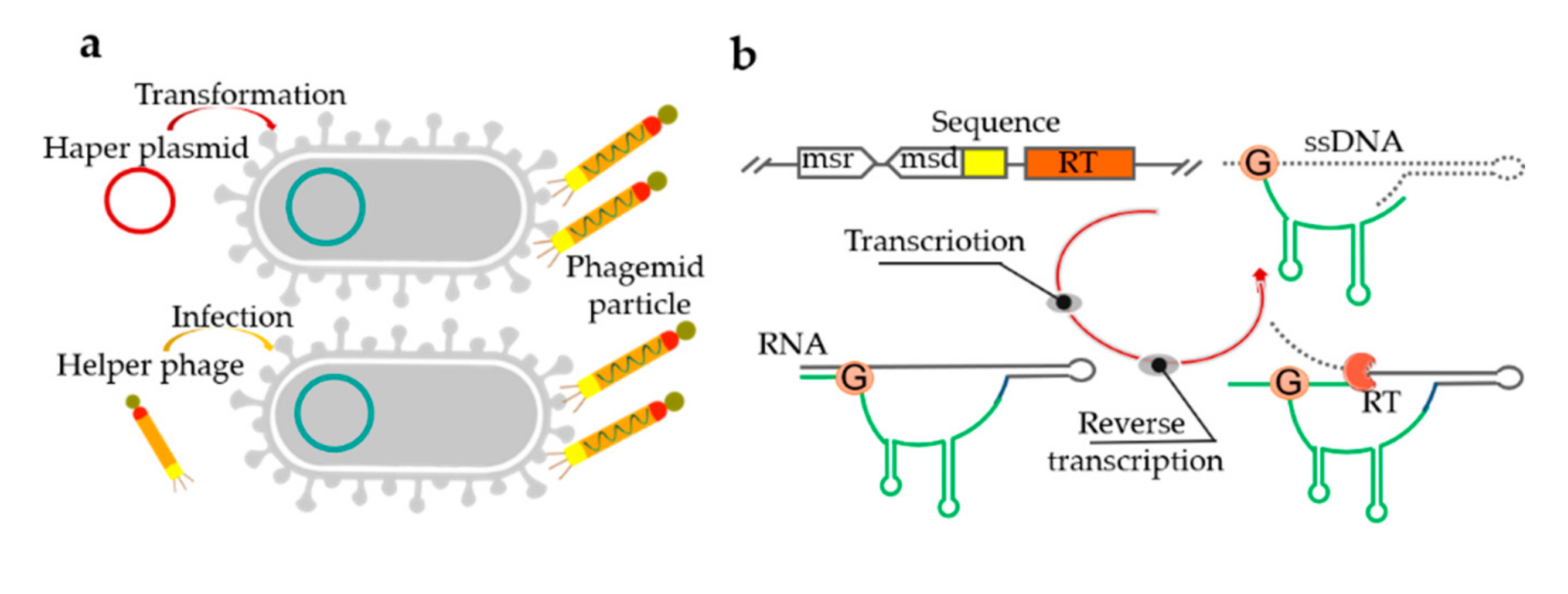
3.2. Transcription and Reverse Transcription
3.3. Asymmetric Polymerase Chain Reaction
3.4. Isothermal Amplification of ssDNA
3.4.1. Primer Exchange Reaction
3.4.2. Rolling Circle Amplification
3.4.3. Other Isothermal Amplification Methods
3.5. Separation of ssDNA from dsDNA
4. Bacteria-Based Production of ssDNA
5. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Farzadfard, F.; Lu, T.K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 2014, 346, 1256272. [Google Scholar] [CrossRef] [Green Version]
- Blackburn, G.M.; Gait, M.J.; Loakes, D.; Williams, D.M.; Egli, M.; Flavell, A.; Allen, S.; Fisher, J.; Haq, S.I.; Engels, J.W. Nucleic Acids in Chemistry and Biology; Royal Society of Chemistry: London, UK, 2006. [Google Scholar]
- Kosuri, S.; Church, G.M. Large-scale de novo DNA synthesis: Technologies and applications. Nat. Methods 2014, 11, 499–507. [Google Scholar] [CrossRef]
- Seguin, J.; Rajeswaran, R.; Malpica-Lopez, N.; Martin, R.R.; Kasschau, K.; Dolja, V.V.; Otten, P.; Farinelli, L.; Pooggin, M.M. De novo reconstruction of consensus master genomes of plant RNA and DNA viruses from siRNAs. PLoS ONE 2014, 9, e88513. [Google Scholar] [CrossRef] [PubMed]
- Giallonardo, F.D.; Töpfer, A.; Rey, M.; Prabhakaran, S.; Duport, Y.; Leemann, C.; Schmutz, S.; Campbell, N.K.; Joos, B.; Lecca, M.R. Full-length haplotype reconstruction to infer the structure of heterogeneous virus populations. Nucleic Acids Res. 2014, 42, e115. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.W.; Chapman, J.; Hugenholtz, P.; Allen, E.E.; Ram, R.J.; Richardson, P.M.; Solovyev, V.V.; Rubin, E.M.; Rokhsar, D.S.; Banfield, J.F. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 2004, 428, 37. [Google Scholar] [CrossRef] [PubMed]
- Caruthers, M.H. Deciphering the protein-DNA recognition code. Acc. Chem. Res. 1980, 13, 155–160. [Google Scholar] [CrossRef]
- Kim, I.; Miller, C.R.; Young, D.L.; Fields, S. High-throughput analysis of in vivo protein stability. Mol. Cell. Proteom. 2013, 12, 3370–3378. [Google Scholar] [CrossRef] [Green Version]
- McLaughlin, R.N., Jr.; Poelwijk, F.J.; Raman, A.; Gosal, W.S.; Ranganathan, R. The spatial architecture of protein function and adaptation. Nature 2012, 491, 138. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, K.A.; McLaughlin, R.N.; Ranganathan, R. Hot spots for allosteric regulation on protein surfaces. Cell 2011, 147, 1564–1575. [Google Scholar] [CrossRef] [Green Version]
- Patwardhan, R.P.; Lee, C.; Litvin, O.; Young, D.L.; Pe’er, D.; Shendure, J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 2009, 27, 1173. [Google Scholar] [CrossRef] [Green Version]
- Schlabach, M.R.; Hu, J.K.; Li, M.; Elledge, S.J. Synthetic design of strong promoters. Proc. Natl. Acad. Sci. USA 2010, 107, 2538–2543. [Google Scholar] [CrossRef] [Green Version]
- Goodman, D.B.; Church, G.M.; Kosuri, S. Causes and effects of N-terminal codon bias in bacterial genes. Science 2013. [Google Scholar] [CrossRef] [Green Version]
- Lai, W.K.; Pugh, B.F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 2017, 18, 548. [Google Scholar] [CrossRef]
- Melnikov, A.; Murugan, A.; Zhang, X.; Tesileanu, T.; Wang, L.; Rogov, P.; Feizi, S.; Gnirke, A.; Callan, C.G., Jr.; Kinney, J.B. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012, 30, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwasnieski, J.C.; Mogno, I.; Myers, C.A.; Corbo, J.C.; Cohen, B.A. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc. Natl. Acad. Sci. USA 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heiat, M.; Ranjbar, R.; Latifi, A.M.; Rasaee, M.J.; Farnoosh, G. Essential strategies to optimize asymmetric PCR conditions as a reliable method to generate large amount of ssDNA aptamers. Biotechnol. Appl. Biochem. 2017, 64, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Pal, S.; Nangreave, J.; Deng, Z.; Liu, Y.; Yan, H. DNA origami with complex curvatures in three-dimensional space. Science 2011, 332, 342–346. [Google Scholar] [CrossRef] [Green Version]
- Amodio, A.; Del Grosso, E.; Troina, A.; Placidi, E.; Ricci, F. Remote Electronic Control of DNA-Based Reactions and Nanostructure Assembly. Nano Lett. 2018, 18, 2918–2923. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297. [Google Scholar] [CrossRef] [Green Version]
- Huai, C.; Li, G.; Yao, R.; Zhang, Y.; Cao, M.; Kong, L.; Jia, C.; Yuan, H.; Chen, H.; Lu, D. Structural insights into DNA cleavage activation of CRISPR-Cas9 system. Nat. Commun. 2017, 8, 1375. [Google Scholar] [CrossRef] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L. Multiplex genome engineering using CRISPR/Cas systems. Science 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yigit, M.V.; Mazumdar, D.; Lu, Y. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv. Drug Deliv. Rev. 2010, 62, 592–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.S.; Zhang, D.Y. Simulation-guided DNA probe design for consistently ultraspecific hybridization. Nat. Chem. 2015, 7, 545. [Google Scholar] [CrossRef] [Green Version]
- Gopinath, A.; Miyazono, E.; Faraon, A.; Rothemund, P.W. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature 2016, 535, 401. [Google Scholar] [CrossRef]
- Bhatia, D.; Arumugam, S.; Nasilowski, M.; Joshi, H.; Wunder, C.; Chambon, V.; Prakash, V.; Grazon, C.; Nadal, B.; Maiti, P.K. Quantum dot-loaded monofunctionalized DNA icosahedra for single-particle tracking of endocytic pathways. Nat. Nanotechnol. 2016, 11, 1112. [Google Scholar] [CrossRef] [Green Version]
- Kuzuya, A.; Sakai, Y.; Yamazaki, T.; Xu, Y.; Komiyama, M. Nanomechanical DNA origami ‘single-molecule beacons’ directly imaged by atomic force microscopy. Nat. Commun. 2011, 2, 449. [Google Scholar] [CrossRef]
- Hughes, R.A.; Ellington, A.D. Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology. CSH Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Zaher, H.; Unrau, P. Methods in Molecular Biology. Oligonucleotide Synthesis: Methods and Applications; Herdewijn, P., Ed.; Humana Press Inc.: New York, NY, USA, 2004; Volume 288, p. 241. [Google Scholar]
- Michelson, A.M.; Todd, A.R. Nuckotides Part XXXII. Xynthesis of a Dithymidine Dinuleotide Containing a 3′: 5′-Internucleotidic Linkuge. J. Chem. Soc. 1955. [Google Scholar] [CrossRef]
- Hall, R.H.; Sir, A.T.; Webb, R.F. Nucleotides Part XLI. Mixed Anhydrides as Intermediates in the Synthesis of Dinucleoside Phosphates. J. Chem. Soc. 1957. [Google Scholar] [CrossRef]
- Gilham, P.T.; Khorana, H.G. Studies on Polynucleotides. I. A New and General Method for the Chemical Synthesis of the C5″-C3″ Internucleotidic Linkage. Syntheses of Deoxyribo-dinucleotides. J. Am. Chem. Soc. 1958, 80, 6212–6222. [Google Scholar] [CrossRef]
- Roy, S.; Caruthers, M. Synthesis of DNA/RNA and their analogs via phosphoramidite and H-phosphonate chemistries. Molecules 2013, 18, 14268–14284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaucage, S.; Caruthers, M.H. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. [Google Scholar] [CrossRef]
- Caruthers, M.H. A Brief Review of DNA and RNA Chemical Synthesis; Portland Press Limited: London, UK, 2011. [Google Scholar]
- Ma, S.; Tang, N.; Tian, J. DNA synthesis, assembly and applications in synthetic biology. Curr. Opin. Chem. Biol. 2012, 16, 260–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeProust, E.M.; Peck, B.J.; Spirin, K.; McCuen, H.B.; Moore, B.; Namsaraev, E.; Caruthers, M.H. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010, 38, 2522–2540. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef]
- Caruthers, M.H. The chemical synthesis of DNA/RNA: Our gift to science. J. Biol. Chem. 2013, 288, 1420–1427. [Google Scholar] [CrossRef] [Green Version]
- Fodor, S.P.; Read, J.L.; Pirrung, M.C.; Stryer, L.; Lu, A.T.; Solas, D. Light-directed, spatially addressable parallel chemical synthesis. Science 1991, 251, 767–773. [Google Scholar] [CrossRef] [Green Version]
- Pease, A.C.; Solas, D.; Sullivan, E.J.; Cronin, M.T.; Holmes, C.P.; Fodor, S. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. USA 1994, 91, 5022–5026. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.Y.; Chen, H.H.; Kao, Y.S.; Kao, W.C.; Peck, K. High throughput parallel synthesis of oligonucleotides with 1536 channel synthesizer. Nucleic Acids Res. 2002, 30, e93. [Google Scholar] [CrossRef] [Green Version]
- Meiser, L.C.; Antkowiak, P.L.; Koch, J.; Chen, W.D.; Kohll, A.X.; Stark, W.J.; Heckel, R.; Grass, R.N. Reading and writing digital data in DNA. Nat. Protoc. 2019. [Google Scholar] [CrossRef] [PubMed]
- Maurer, K.; Cooper, J.; Caraballo, M.; Crye, J.; Suciu, D.; Ghindilis, A.; Leonetti, J.A.; Wang, W.; Rossi, F.M.; Stöver, A.G. Electrochemically generated acid and its containment to 100 micron reaction areas for the production of DNA microarrays. PLoS ONE 2006, 1, e34. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Gulari, E.; Zhou, X. In situ synthesis of oligonucleotide microarrays. Biopolymers 2004, 73, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Organick, L.; Ang, S.D.; Chen, Y.-J.; Lopez, R.; Yekhanin, S.; Makarychev, K.; Racz, M.Z.; Kamath, G.; Gopalan, P.; Nguyen, B. Random access in large-scale DNA data storage. Nat. Biotechnol. 2018, 36, 242. [Google Scholar] [CrossRef] [PubMed]
- Beaucage, S.L. Strategies in the preparation of DNA oligonucleotide arrays for diagnostic applications. Curr. Med. Chem. 2001, 8, 1213–1244. [Google Scholar] [CrossRef]
- Abramova, T. Frontiers and approaches to chemical synthesis of oligodeoxyribonucleotides. Molecules 2013, 18, 1063–1075. [Google Scholar] [CrossRef]
- Wan, W.; Wang, D.; Gao, X.; Hong, J. Immobilized MutS-Mediated Error Removal of Microchip-Synthesized DNA. In Synthetic DNA; Springer: New York, NY, USA, 2017; pp. 217–235. [Google Scholar]
- Wan, W.; Li, L.; Xu, Q.; Wang, Z.; Yao, Y.; Wang, R.; Zhang, J.; Liu, H.; Gao, X.; Hong, J. Error removal in microchip-synthesized DNA using immobilized MutS. Nucleic Acids Res. 2014, 42, e102. [Google Scholar] [CrossRef] [Green Version]
- Loc’h, J.; Delarue, M. Terminal deoxynucleotidyltransferase: The story of an untemplated DNA polymerase capable of DNA bridging and templated synthesis across strands. Curr. Opin. Struct. Biol. 2018, 53, 22–31. [Google Scholar] [CrossRef]
- Palluk, S.; Arlow, D.H.; de Rond, T.; Barthel, S.; Kang, J.S.; Bector, R.; Baghdassarian, H.M.; Truong, A.N.; Kim, P.W.; Singh, A.K.; et al. De novo DNA synthesis using polymerase-nucleotide conjugates. Nat. Biotechnol. 2018, 36, 645–650. [Google Scholar] [CrossRef]
- Miura, H.; Quadros, R.M.; Gurumurthy, C.B.; Ohtsuka, M. Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat. Protoc. 2018, 13, 195. [Google Scholar] [CrossRef]
- Xie, X.; Yang, R. Multi-copy single-stranded DNA in Escherichia coli. Microbiology 2017, 163, 1735–1739. [Google Scholar] [CrossRef] [PubMed]
- Veneziano, R.; Shepherd, T.R.; Ratanalert, S.; Bellou, L.; Tao, C.; Bathe, M. In vitro synthesis of gene-length single-stranded DNA. Sci. Rep. 2018, 8, 6548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollenstein, M. DNA Synthesis by Primer Exchange Reaction Cascades. ChemBioChem 2018, 19, 422–424. [Google Scholar] [CrossRef] [PubMed]
- Kishi, J.Y.; Schaus, T.E.; Gopalkrishnan, N.; Xuan, F.; Yin, P. Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. 2018, 10, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Johne, R.; Müller, H.; Rector, A.; Van Ranst, M.; Stevens, H. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 2009, 17, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Joneja, A.; Huang, X. Linear nicking endonuclease-mediated strand-displacement DNA amplification. Anal. Biochem. 2011, 414, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Motea, E.A.; Berdis, A.J. Terminal deoxynucleotidyl transferase: The story of a misguided DNA polymerase. BBA-Proteins Proteom. 2010, 1804, 1151–1166. [Google Scholar] [CrossRef] [Green Version]
- Fowler, J.D.; Suo, Z. Biochemical, structural, and physiological characterization of terminal deoxynucleotidyl transferase. Chem. Rev. 2006, 106, 2092–2110. [Google Scholar] [CrossRef]
- Jensen, M.A.; Davis, R.W. Template-independent enzymatic oligonucleotide synthesis (TiEOS): Its history, prospects, and challenges. Biochemistry 2018, 57, 1821–1832. [Google Scholar] [CrossRef]
- Jang, E.K.; Son, R.G.; Pack, S.P. Novel enzymatic single-nucleotide modification of DNA oligomer: Prevention of incessant incorporation of nucleotidyl transferase by ribonucleotide-borate complex. Nucleic Acids Res. 2019, 47, e102. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Liu, Z.; Li, W.; Huang, Y.; Nie, Z.; Yao, S. Enzymatically generated long polyT-templated copper nanoparticles for versatile biosensing assay of DNA-related enzyme activity. Anal. Methods 2015, 7, 4355–4361. [Google Scholar] [CrossRef]
- Tang, L.; Navarro, L.A., Jr.; Chilkoti, A.; Zauscher, S. High-Molecular-Weight Polynucleotides by Transferase-Catalyzed Living Chain-Growth Polycondensation. Angewandte Chemie 2017, 56, 6778–6782. [Google Scholar] [CrossRef]
- Lee, H.H.; Kalhor, R.; Goela, N.; Bolot, J.; Church, G.M. Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nat. Commun. 2019, 10, 2383. [Google Scholar] [CrossRef] [Green Version]
- Jie, G.; Ge, J.; Gao, X.; Li, C. Amplified electrochemiluminescence detection of CEA based on magnetic Fe3O4@ Au nanoparticles-assembled Ru@ SiO2 nanocomposites combined with multiple cycling amplification strategy. Biosens. Bioelectron. 2018, 118, 115–121. [Google Scholar] [CrossRef]
- Miura, H.; Gurumurthy, C.B.; Sato, T.; Sato, M.; Ohtsuka, M. CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Sci. Rep. 2015, 5, 12799. [Google Scholar] [CrossRef] [Green Version]
- Codner, G.F.; Mianné, J.; Caulder, A.; Loeffler, J.; Fell, R.; King, R.; Allan, A.J.; Mackenzie, M.; Pike, F.J.; McCabe, C.V. Application of long single-stranded DNA donors in genome editing: Generation and validation of mouse mutants. BMC Biol. 2018, 16, 70. [Google Scholar] [CrossRef]
- Tolnai, Z.; Harkai, Á.; Szeitner, Z.; Scholz, É.N.; Percze, K.; Gyurkovics, A.; Mészáros, T. A simple modification increases specificity and efficiency of asymmetric PCR. Anal. Chim. Acta 2019, 1047, 225–230. [Google Scholar] [CrossRef]
- Gyllensten, U.B.; Erlich, H.A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc. Natl. Acad. Sci. USA 1988, 85, 7652–7656. [Google Scholar] [CrossRef] [Green Version]
- Marimuthu, C.; Tang, T.H.; Tominaga, J.; Tan, S.C.; Gopinath, S.C. Single-stranded DNA (ssDNA) production in DNA aptamer generation. Analyst 2012, 137, 1307–1315. [Google Scholar] [CrossRef]
- Ding, J.; Gan, S.; Ho, B. Single-stranded DNA oligoaptamers: Molecular recognition and LPS antagonism are length-and secondary structure-dependent. J. Innate Immun. 2009, 1, 46–58. [Google Scholar] [CrossRef]
- Wooddell, C.I.; Burgess, R.R. Use of asymmetric PCR to generate long primers and single-stranded DNA for incorporating cross-linking analogs into specific sites in a DNA probe. Genome Res. 1996, 6, 886–892. [Google Scholar] [CrossRef] [Green Version]
- Veneziano, R.; Ratanalert, S.; Zhang, K.; Zhang, F.; Yan, H.; Chiu, W.; Bathe, M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. [Google Scholar] [CrossRef] [Green Version]
- Tolle, F.; Wilke, J.; Wengel, J.; Mayer, G. By-product formation in repetitive PCR amplification of DNA libraries during SELEX. PLoS ONE 2014, 9, e114693. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Tanner, N.A. Isothermal amplification of long, discrete DNA fragments facilitated by single-stranded binding protein. Sci. Rep. 2017, 7, 8497. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal amplification of nucleic acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Baccouche, A.; Montagne, K.; Padirac, A.; Fujii, T.; Rondelez, Y. Dynamic DNA-toolbox reaction circuits: A walkthrough. Methods 2014, 67, 234–249. [Google Scholar] [CrossRef]
- Minev, D.; Guerra, R.; Kishi, J.; Smith, C.; Krieg, E.; Said, K.; Hornick, A.; Sasaki, H.; Filsinger, G.; Beliveau, B. Rapid and scalable in vitro production of single-stranded DNA. BioRxiv 2019. [Google Scholar] [CrossRef]
- Dean, F.B.; Nelson, J.R.; Giesler, T.L.; Lasken, R.S. Rapid amplification of plasmid and phage DNA using phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001, 11, 1095–1099. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.; Yan, W.; Liu, L.; Wang, S.; Zhang, X.; Lyu, M. Research progress on rolling circle amplification (RCA)-based biomedical sensing. Pharmaceuticals 2018, 11, 35. [Google Scholar] [CrossRef] [Green Version]
- Hollenstein, M. Generation of long, fully modified, and serum-resistant oligonucleotides by rolling circle amplification. Org. Biomol. Chem. 2015, 13, 9820–9824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducani, C.; Kaul, C.; Moche, M.; Shih, W.M.; Högberg, B. Enzymatic production of’monoclonal stoichiometric’single-stranded DNA oligonucleotides. Nat. Methods 2013, 10, 647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelissen, F.H.; Goossens, E.P.; Tessari, M.; Heus, H.A. Enzymatic preparation of multimilligram amounts of pure single-stranded DNA samples for material and analytical sciences. Anal. Biochem. 2015, 475, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Van Emmerik, C.L.; Gachulincova, I.; Lobbia, V.R.; Daniëls, M.A.; Heus, H.A.; Soufi, A.; Nelissen, F.H.; van Ingen, H. Ramified rolling circle amplification for synthesis of nucleosomal DNA sequences. Anal. Biochem. 2020, 588, 113469. [Google Scholar] [CrossRef]
- Shi, D.; Huang, J.; Chuai, Z.; Chen, D.; Zhu, X.; Wang, H.; Peng, J.; Wu, H.; Huang, Q.; Fu, W. Isothermal and rapid detection of pathogenic microorganisms using a nano-rolling circle amplification-surface plasmon resonance biosensor. Biosens. Bioelectron. 2014, 62, 280–287. [Google Scholar] [CrossRef]
- Huang, R.; Li, S.; Liu, H.; Jin, L.; Zhao, Y.; Li, Z.; He, N. A simple fluorescence aptasensor for gastric cancer exosome detection based on branched rolling circle amplification. Nanoscale 2019. [Google Scholar] [CrossRef]
- Kim, T.-Y.; Lim, M.-C.; Woo, M.-A.; Jun, B.-H. Radial flow assay using gold nanoparticles and rolling circle amplification to detect mercuric ions. Nanomaterials 2018, 8, 81. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Li, L.; Duan, L.; Wang, X.; Xie, Y.; Tong, L.; Wang, Q.; Tang, B. High specific and ultrasensitive isothermal detection of microRNA by padlock probe-based exponential rolling circle amplification. Anal. Chem. 2013, 85, 7941–7947. [Google Scholar] [CrossRef]
- Demidov, V.V. Rolling-circle amplification in DNA diagnostics: The power of simplicity. Expert Rev. Mol. Diagn. 2002, 2, 542–548. [Google Scholar] [CrossRef]
- Zyrina, N.V.; Zheleznaya, L.A.; Dvoretsky, E.V.; Vasiliev, V.D.; Chernov, A.; Matvienko, N.I. BspD6I DNA nickase strongly stimulates template-independent synthesis of non-palindromic repetitive DNA by Bst DNA polymerase. Biol. Chem. 2007, 388, 367–372. [Google Scholar] [CrossRef]
- Cui, W.; Wang, L.; Xu, X.; Wang, Y.; Jiang, W. A loop-mediated cascade amplification strategy for highly sensitive detection of DNA methyltransferase activity. Sens. Actuators B Chem. 2017, 244, 599–605. [Google Scholar] [CrossRef]
- Meijer, L.H.; Joesaar, A.; Steur, E.; Engelen, W.; Van Santen, R.A.; Merkx, M.; De Greef, T.F. Hierarchical control of enzymatic actuators using DNA-based switchable memories. Nat. Commun. 2017, 8, 1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuffrida, M.C.; Spoto, G. Integration of isothermal amplification methods in microfluidic devices: Recent advances. Biosens. Bioelectron. 2017, 90, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Gui, G.-F.; Zhuo, Y.; Chai, Y.-Q.; Xiang, Y.; Yuan, R. Signal-off electrochemiluminescence biosensor based on Phi29 DNA polymerase mediated strand displacement amplification for microRNA detection. Anal. Chem. 2015, 87, 6328–6334. [Google Scholar] [CrossRef]
- Fakruddin, M.; Mannan, K.S.B.; Chowdhury, A.; Mazumdar, R.M.; Hossain, M.N.; Islam, S.; Chowdhury, M.A. Nucleic acid amplification: Alternative methods of polymerase chain reaction. J. Pharm. Bioallied Sci. 2013, 5, 245. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [Green Version]
- Abdullahi, U.F.; Naim, R.; Taib, W.R.W.; Saleh, A.; Muazu, A.; Aliyu, S.; Baig, A.A. Loop-mediated isothermal amplification (LAMP), an innovation in gene amplification: Bridging the gap in molecular diagnostics; a review. Indian J. Sci. Technol. 2015, 8, 1. [Google Scholar] [CrossRef]
- Nagamine, K.; Kuzuhara, Y.; Notomi, T. Isolation of single-stranded DNA from loop-mediated isothermal amplification products. Biochem. Biophys. Res. Commun. 2002, 290, 1195–1198. [Google Scholar] [CrossRef]
- Damase, T.R.; Ellington, A.D.; Allen, P.B. Purification of single-stranded DNA by co-polymerization with acrylamide and electrophoresis. BioTechniques 2017, 62, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Pagratis, N.C. Rapid preparation of single stranded DNA from PCR products by streptavidin induced electrophoretic mobility shift. Nucleic Acids Res. 1996, 24, 3645–3646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Chao, J.; Pan, D.; Liu, H.; Huang, Q.; Fan, C. Folding super-sized DNA origami with scaffold strands from long-range PCR. Chem. Commun. 2012. [Google Scholar] [CrossRef] [PubMed]
- Erkelenz, M.; Bauer, D.M.; Meyer, R.; Gatsogiannis, C.; Raunser, S.; Sacca, B.; Niemeyer, C.M. A facile method for preparation of tailored scaffolds for DNA-origami. Small 2014, 10, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; McGown, L.B. Sequence-based separation of single-stranded DNA at high salt concentrations in capillary zone electrophoresis. Electrophoresis 2016, 37, 2017–2024. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R. Preparation of single-stranded DNA from PCR products with streptavidin magnetic beads. Nucleic Acid Ther. 2011, 21, 437–440. [Google Scholar] [CrossRef]
- Kilili, G.K.; Tilton, L.; Karbiwnyk, C.M. [Letter to the Editor] NaOH concentration and streptavidin bead type are key factors for optimal DNA aptamer strand separation and isolation. BioTechniques 2016, 61, 114–116. [Google Scholar] [CrossRef] [Green Version]
- Kao, W.S.; Li, H.W. An Efficient Bead-captured Denaturation Method for Preparing Long Single-stranded DNA. J. Chin. Chem. Soc. 2017, 64, 1065–1070. [Google Scholar] [CrossRef]
- Holmberg, A.; Blomstergren, A.; Nord, O.; Lukacs, M.; Lundeberg, J.; Uhlén, M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis 2005, 26, 501–510. [Google Scholar] [CrossRef]
- Kujau, M.J.; Wölfl, S. Efficient preparation of single-stranded DNA for in vitro selection. Mol. Biotechnol. 1997, 7, 333–335. [Google Scholar] [CrossRef]
- Engelhardt, F.A.; Praetorius, F.; Wachauf, C.H.; Brüggenthies, G.; Kohler, F.; Kick, B.; Kadletz, K.L.; Pham, P.N.; Behler, K.L.; Gerling, T. Custom-Size, Functional, and Durable DNA Origami with Design-Specific Scaffolds. ACS Nano 2019. [Google Scholar] [CrossRef]
- Kick, B.; Praetorius, F.; Dietz, H.; Weuster-Botz, D. Efficient production of single-stranded phage DNA as scaffolds for DNA origami. Nano Lett. 2015, 15, 4672–4676. [Google Scholar] [CrossRef] [PubMed]
- Kick, B.; Hensler, S.; Praetorius, F.; Dietz, H.; Weuster-Botz, D. Specific growth rate and multiplicity of infection affect high-cell-density fermentation with bacteriophage M13 for ssDNA production. Biotechnol. Bioeng. 2017, 114, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Marchi, A.N.; Saaem, I.; Vogen, B.N.; Brown, S.; LaBean, T.H. Toward larger DNA origami. Nano Lett. 2014, 14, 5740–5747. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Dong, Q.; Ma, R.; Chen, Y.; Yang, J.; Sun, L.-Z.; Huang, C. Rapid isolation of highly pure single-stranded DNA from phagemids. Anal. Biochem. 2009, 389, 177–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nafisi, P.M.; Aksel, T.; Douglas, S.M. Construction of a novel phagemid to produce custom DNA origami scaffolds. Synth. Biol. 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Chasteen, L.; Ayriss, J.; Pavlik, P.; Bradbury, A. Eliminating helper phage from phage display. Nucleic Acids Res. 2006, 34, e145. [Google Scholar] [CrossRef] [Green Version]
- Praetorius, F.; Kick, B.; Behler, K.L.; Honemann, M.N.; Weuster-Botz, D.; Dietz, H. Biotechnological mass production of DNA origami. Nature 2017, 552, 84–87. [Google Scholar] [CrossRef]
- Lim, D.; Maas, W.K. Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA-RNA compound in E. coli B. Cell 1989, 56, 891–904. [Google Scholar] [CrossRef]
- Shimada, M.; Inouye, S.; Inouye, M. Requirements of the secondary structures in the primary transcript for multicopy single-stranded DNA synthesis by reverse transcriptase from bacterial retron-Ec107. J. Biol. Chem. 1994, 269, 14553–14558. [Google Scholar] [CrossRef]
- Elbaz, J.; Yin, P.; Voigt, C.A. Genetic encoding of DNA nanostructures and their self-assembly in living bacteria. Nat. Commun. 2016, 7, 11179. [Google Scholar] [CrossRef]
- Riele, H.T.; Michel, B.; Ehrlich, S.D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986, 5, 631. [Google Scholar] [CrossRef]
- Bruand, C.; Ehrlich, S.D. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol. Microbiol. 2000, 35. [Google Scholar] [CrossRef] [PubMed]



| Strategy | Template | Enzymes | Product Separation | Primer Design | Single Step Technique | Refs. |
|---|---|---|---|---|---|---|
| TdT | No | Terminal deoxynucleotide transferase | No | No | No | [52,53] |
| ivTRT | Yes | RNA polymerase, reverse transcriptase and RNaseH | Yes | Simple | No | [54,55] |
| aPCR | Yes | DNA polymerase | Yes | Simple | Yes | [56] |
| PER | Yes | DNA polymerase | Yes | Complex | Yes | [57,58] |
| RCA | Yes | DNA polymerase | Yes | Complex | Yes | [59,60] |
| SDA | Yes | DNA polymerase and strand-limited restriction endonuclease/nicking enzym | Yes | Complex | Yes | [61] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, M.; Qiao, J.; Qi, H. Current and Emerging Methods for the Synthesis of Single-Stranded DNA. Genes 2020, 11, 116. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11020116
Hao M, Qiao J, Qi H. Current and Emerging Methods for the Synthesis of Single-Stranded DNA. Genes. 2020; 11(2):116. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11020116
Chicago/Turabian StyleHao, Min, Jianjun Qiao, and Hao Qi. 2020. "Current and Emerging Methods for the Synthesis of Single-Stranded DNA" Genes 11, no. 2: 116. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11020116