Embryo-Based Large Fragment Knock-in in Mammals: Why, How and What’s Next
Abstract
:1. Introduction
2. Why Large Fragment Knock-In?
2.1. Let There Be Light—Developmental Biology in Real-Time
2.2. Understanding Our Past—Functional Evolution
2.3. Mouse or Human – Genetic Humanization in Medical Research
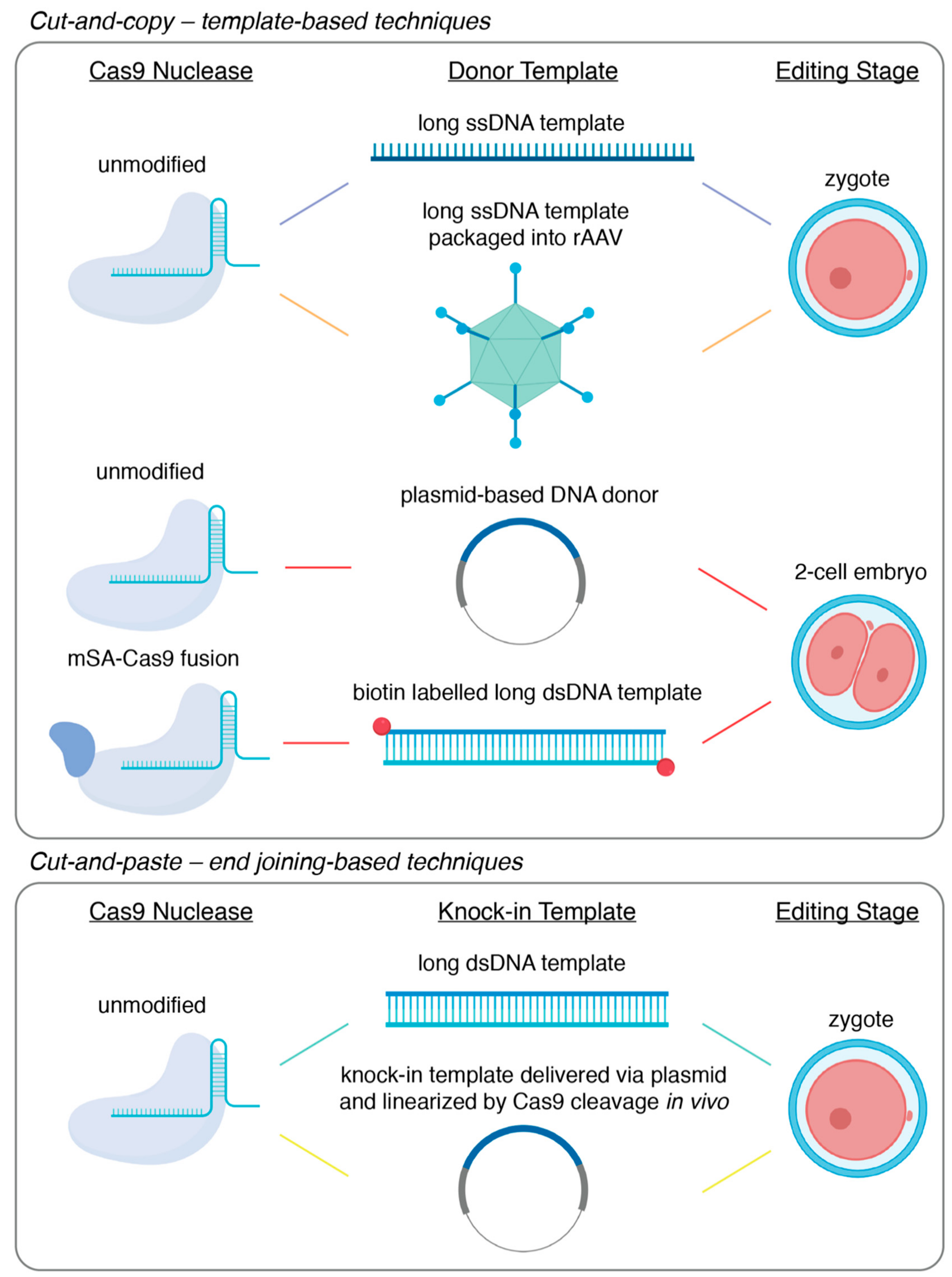
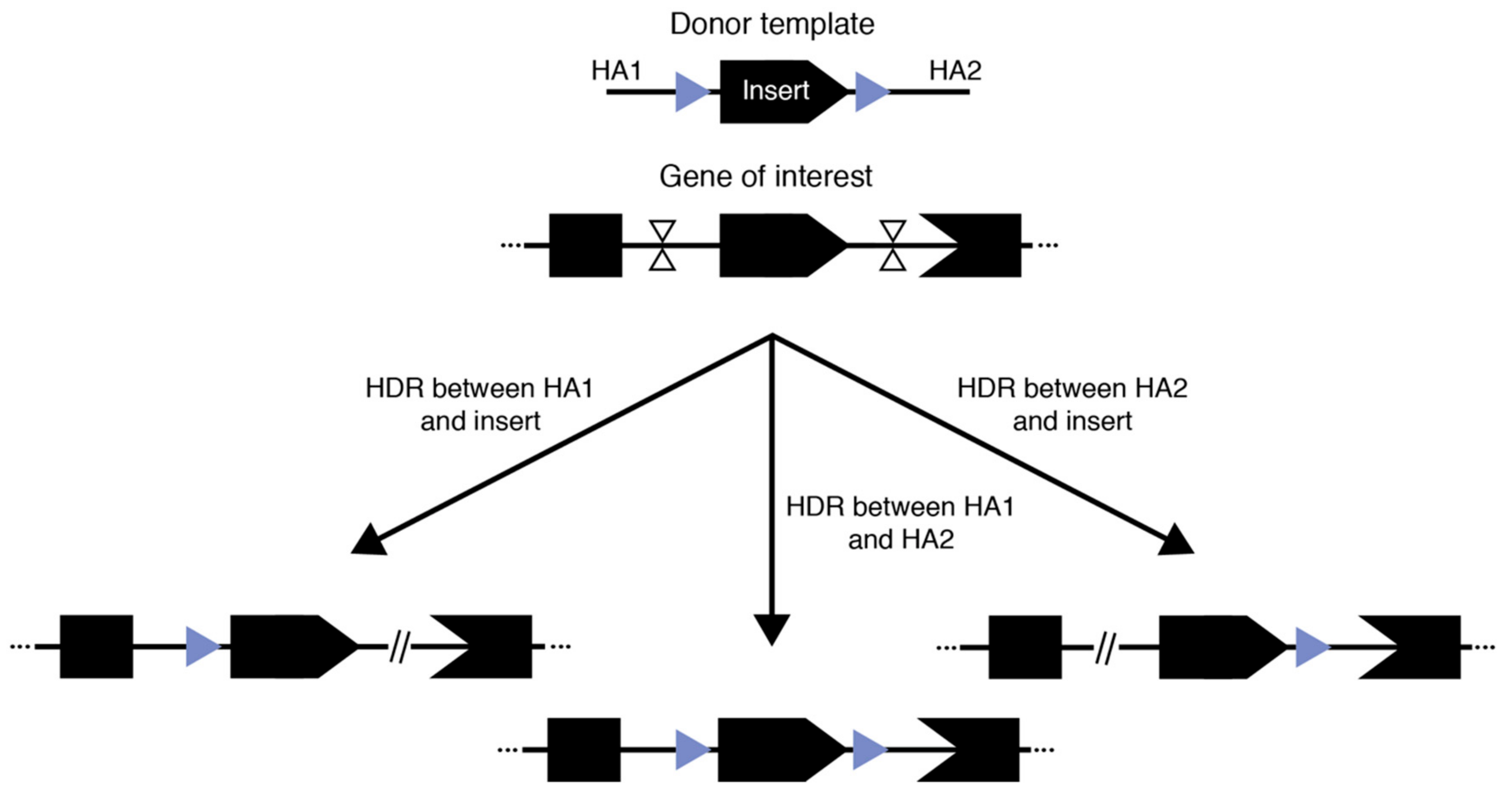
3. How to Achieve Large Fragment Knock-In
3.1. Cut-and-Copy—Template-Based Techniques
3.2. Cut-and-Paste – End Joining-Based Techniques
3.3. Conditional Alleles—What is the Better Way
4. Future—Large and Larger
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertolini, L.R.; Meade, H.; Lazzarotto, C.R.; Martins, L.T.; Tavares, K.C.; Bertolini, M.; Murray, J.D. The transgenic animal platform for biopharmaceutical production. Transgenic. Res. 2016, 25, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Chu, C.; Wang, F.; Niu, Y. CRISPR/Cas9-mediated genome editing in nonhuman primates. Dis. Model Mech. 2019, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Bindels, D.S.; Haarbosch, L.; van Weeren, L.; Postma, M.; Wiese, K.E.; Mastop, M.; Aumonier, S.; Gotthard, G.; Royant, A.; Hink, M.A.; et al. mScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 2017, 14, 53–56. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Baloban, M.; Verkhusha, V.V. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr. Opin. Chem. Biol. 2015, 27, 52–63. [Google Scholar] [CrossRef]
- Los, G.V.; Encell, L.P.; McDougall, M.G.; Hartzell, D.D.; Karassina, N.; Zimprich, C.; Wood, M.G.; Learish, R.; Ohana, R.F.; Urh, M.; et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008, 3, 373–382. [Google Scholar] [CrossRef]
- Xu, T.; Close, D.; Handagama, W.; Marr, E.; Sayler, G.; Ripp, S. The expanding toolbox of in vivo bioluminescent imaging. Front. Oncol. 2016, 6, 150. [Google Scholar] [CrossRef] [Green Version]
- Kolberg, K.; Puettmann, C.; Pardo, A.; Fitting, J.; Barth, S. SNAP-tag technology: A general introduction. Curr. Pharm. Des. 2013, 19, 5406–5413. [Google Scholar] [CrossRef]
- Knight, S.C.; Tjian, R.; Doudna, J.A. Genomes in focus: Development and applications of CRISPR-Cas9 imaging technologies. Angew. Chem. Int. Ed. Engl. 2018, 57, 4329–4337. [Google Scholar] [CrossRef]
- Rodriguez, A.J.; Condeelis, J.; Singer, R.H.; Dictenberg, J.B. Imaging mRNA movement from transcription sites to translation sites. Semin. Cell Dev. Biol. 2007, 18, 202–208. [Google Scholar] [CrossRef] [Green Version]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boersma, S.; Khuperkar, D.; Verhagen, B.M.P.; Sonneveld, S.; Grimm, J.B.; Lavis, L.D.; Tanenbaum, M.E. Multi-Color Single-Molecule Imaging Uncovers Extensive Heterogeneity in mRNA Decoding. Cell 2019, 178, 458–472.e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benninger, R.K.; Hao, M.; Piston, D.W. Multi-photon excitation imaging of dynamic processes in living cells and tissues. Rev. Physiol. Biochem. Pharmacol. 2008, 160, 71–92. [Google Scholar] [PubMed]
- Wan, Y.; McDole, K.; Keller, P.J. Light-Sheet Microscopy and Its Potential for Understanding Developmental Processes. Annu. Rev. Cell Dev. Biol. 2019, 35, 655–681. [Google Scholar] [CrossRef] [Green Version]
- Keller, P.J.; Schmidt, A.D.; Wittbrodt, J.; Stelzer, E.H. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 2008, 322, 1065–1069. [Google Scholar] [CrossRef] [Green Version]
- Amat, F.; Keller, P.J. Towards comprehensive cell lineage reconstructions in complex organisms using light-sheet microscopy. Dev Growth Differ 2013, 55, 563–578. [Google Scholar] [CrossRef]
- Wan, Y.; Wei, Z.; Looger, L.L.; Koyama, M.; Druckmann, S.; Keller, P.J. Single-Cell Reconstruction of Emerging Population Activity in an Entire Developing Circuit. Cell 2019, 179, 355–372.e23. [Google Scholar] [CrossRef]
- McDole, K.; Guignard, L.; Amat, F.; Berger, A.; Malandain, G.; Royer, L.A.; Turaga, S.C.; Branson, K.; Keller, P.J. In toto imaging and reconstruction of post-implantation mouse development at the single-cell level. Cell 2018, 175, 859–876.e33. [Google Scholar] [CrossRef] [Green Version]
- Amat, F.; Hockendorf, B.; Wan, Y.; Lemon, W.C.; McDole, K.; Keller, P.J. Efficient processing and analysis of large-scale light-sheet microscopy data. Nat. Protoc. 2015, 10, 1679–1696. [Google Scholar] [CrossRef]
- Ikawa, M.; Yamada, S.; Nakanishi, T.; Okabe, M. ‘Green mice’ and their potential usage in biological research. FEBS Lett. 1998, 430, 83–87. [Google Scholar] [CrossRef]
- Palmiter, R.D.; Norstedt, G.; Gelinas, R.E.; Hammer, R.E.; Brinster, R.L. Metallothionein-human GH fusion genes stimulate growth of mice. Science 1983, 222, 809–814. [Google Scholar] [CrossRef] [PubMed]
- Laboulaye, M.A.; Duan, X.; Qiao, M.; Whitney, I.E.; Sanes, J.R. Mapping Transgene Insertion Sites Reveals Complex Interactions Between Mouse Transgenes and Neighboring Endogenous Genes. Front. Mol. Neurosci. 2018, 11, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weis, J.; Fine, S.M.; Sanes, J.R. Integration site-dependent transgene expression used to mark subpopulations of cells in vivo: An example from the neuromuscular junction. Brain Pathol. 1992, 2, 31–37. [Google Scholar]
- Goodwin, L.O.; Splinter, E.; Davis, T.L.; Urban, R.; He, H.; Braun, R.E.; Chesler, E.J.; Kumar, V.; van Min, M.; Ndukum, J.; et al. Large-scale discovery of mouse transgenic integration sites reveals frequent structural variation and insertional mutagenesis. Genome Res. 2019, 29, 494–505. [Google Scholar] [CrossRef] [Green Version]
- Thomas, K.R.; Capecchi, M.R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 1987, 51, 503–512. [Google Scholar] [CrossRef]
- Capecchi, M.R. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005, 6, 507–512. [Google Scholar] [CrossRef]
- Tan, W.; Proudfoot, C.; Lillico, S.G.; Whitelaw, C.B. Gene targeting, genome editing: From Dolly to editors. Transgenic. Res. 2016, 25, 273–287. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J. ‘Any idiot can do it’. Genome editor CRISPR could put mutant mice in everyone’s reach. Science 2016. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.H. Molecular evolution meets the genomics revolution. Nat. Genet. 2003, 33, 255–265. [Google Scholar] [CrossRef]
- Ellegren, H. Evolution: Natural selection in the evolution of humans and chimps. Curr. Biol. 2005, 15, R919–R922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reilly, S.K.; Noonan, J.P. Evolution of Gene Regulation in Humans. Annu. Rev. Genomics Hum. Genet. 2016, 17, 45–67. [Google Scholar] [CrossRef] [PubMed]
- Behringer, R.R.; Rasweiler, J.J.T.; Chen, C.H.; Cretekos, C.J. Genetic regulation of mammalian diversity. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, S.A.; Glimm, T.; Bhat, R. The vertebrate limb: An evolving complex of self-organizing systems. Prog. Biophys. Mol. Biol. 2018, 137, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Kvon, E.Z.; Kamneva, O.K.; Melo, U.S.; Barozzi, I.; Osterwalder, M.; Mannion, B.J.; Tissieres, V.; Pickle, C.S.; Plajzer-Frick, I.; Lee, E.A.; et al. Progressive Loss of Function in a Limb Enhancer during Snake Evolution. Cell 2016, 167, 633–642.e11. [Google Scholar] [CrossRef] [Green Version]
- Enard, W.; Przeworski, M.; Fisher, S.E.; Lai, C.S.; Wiebe, V.; Kitano, T.; Monaco, A.P.; Paabo, S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 2002, 418, 869–872. [Google Scholar] [CrossRef]
- Enard, W.; Gehre, S.; Hammerschmidt, K.; Holter, S.M.; Blass, T.; Somel, M.; Bruckner, M.K.; Schreiweis, C.; Winter, C.; Sohr, R.; et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 2009, 137, 961–971. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, B. Sydney Brenner—A personal perspective. Genome Res. 2019, 29, vii-ix. [Google Scholar] [CrossRef]
- Aparicio, S.; Chapman, J.; Stupka, E.; Putnam, N.; Chia, J.M.; Dehal, P.; Christoffels, A.; Rash, S.; Hoon, S.; Smit, A.; et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 2002, 297, 1301–1310. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, B.; Si-Hoe, S.L.; Murphy, D.; Brenner, S. Transgenic rats reveal functional conservation of regulatory controls between the Fugu isotocin and rat oxytocin genes. Proc. Natl. Acad. Sci. USA 1997, 94, 12462–12466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.S.; Raes, J. Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 2004, 38, 615–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adikusuma, F.; Pederick, D.; McAninch, D.; Hughes, J.; Thomas, P. Functional Equivalence of the SOX2 and SOX3 Transcription Factors in the Developing Mouse Brain and Testes. Genetics 2017, 206, 1495–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, F.; Nair, R.R.; Fisher, E.M.C.; Cunningham, T.J. Humanising the mouse genome piece by piece. Nat. Commun. 2019, 10, 1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douam, F.; Gaska, J.M.; Winer, B.Y.; Ding, Q.; von Schaewen, M.; Ploss, A. Genetic Dissection of the Host Tropism of Human-Tropic Pathogens. Annu. Rev. Genet. 2015, 49, 21–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorner, M.; Horwitz, J.A.; Donovan, B.M.; Labitt, R.N.; Budell, W.C.; Friling, T.; Vogt, A.; Catanese, M.T.; Satoh, T.; Kawai, T.; et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature 2013, 501, 237–241. [Google Scholar] [CrossRef] [Green Version]
- Ding, Q.; von Schaewen, M.; Hrebikova, G.; Heller, B.; Sandmann, L.; Plaas, M.; Ploss, A. Mice expressing minimally humanized CD81 and occludin genes support hepatitis C virus uptake in vivo. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Maeder, M.L.; Stefanidakis, M.; Wilson, C.J.; Baral, R.; Barrera, L.A.; Bounoutas, G.S.; Bumcrot, D.; Chao, H.; Ciulla, D.M.; DaSilva, J.A.; et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019, 25, 229–233. [Google Scholar] [CrossRef]
- Tait-Burkard, C.; Doeschl-Wilson, A.; McGrew, M.J.; Archibald, A.L.; Sang, H.M.; Houston, R.D.; Whitelaw, C.B.; Watson, M. Livestock 2.0—Genome editing for fitter, healthier, and more productive farmed animals. Genome Biol. 2018, 19, 204. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Zhang, J.; Wu, H.; Wang, J.; Ma, K.; Li, Z.; Zhang, X.; Zhang, P.; Huang, X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013, 23, 720–723. [Google Scholar] [CrossRef]
- Inui, M.; Miyado, M.; Igarashi, M.; Tamano, M.; Kubo, A.; Yamashita, S.; Asahara, H.; Fukami, M.; Takada, S. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci. Rep. 2014, 4, 5396. [Google Scholar] [CrossRef] [PubMed]
- Miura, H.; Quadros, R.M.; Gurumurthy, C.B.; Ohtsuka, M. Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat. Protoc. 2018, 13, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Quadros, R.M.; Miura, H.; Harms, D.W.; Akatsuka, H.; Sato, T.; Aida, T.; Redder, R.; Richardson, G.P.; Inagaki, Y.; Sakai, D.; et al. Easi-CRISPR: A robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017, 18, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, B.; Posfai, E.; Rossant, J. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat. Biotechnol. 2018, 36, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sun, S.; Moonen, D.; Lee, C.; Lee, A.Y.; Schaffer, D.V.; He, L. CRISPR-READI: Efficient Generation of Knockin Mice by CRISPR RNP Electroporation and AAV Donor Infection. Cell Rep. 2019, 27, 3780–3789.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Zhang, M.; Wang, X.; Ying, W.; Hu, X.; Dai, P.; Meng, F.; Shi, L.; Sun, Y.; Yao, N.; et al. Tild-CRISPR Allows for Efficient and Precise Gene Knockin in Mouse and Human Cells. Dev. Cell 2018, 45, 526–536.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakade, S.; Tsubota, T.; Sakane, Y.; Kume, S.; Sakamoto, N.; Obara, M.; Daimon, T.; Sezutsu, H.; Yamamoto, T.; Sakuma, T.; et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014, 5, 5560. [Google Scholar] [CrossRef]
- Aida, T.; Nakade, S.; Sakuma, T.; Izu, Y.; Oishi, A.; Mochida, K.; Ishikubo, H.; Usami, T.; Aizawa, H.; Yamamoto, T.; et al. Gene cassette knock-in in mammalian cells and zygotes by enhanced MMEJ. BMC Genomics 2016, 17, 979. [Google Scholar] [CrossRef] [Green Version]
- Gurumurthy, C.B.; O’Brien, A.R.; Quadros, R.M.; Adams, J., Jr.; Alcaide, P.; Ayabe, S.; Ballard, J.; Batra, S.K.; Beauchamp, M.C.; Becker, K.A.; et al. Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: A multi-center evaluation. Genome Biol. 2019, 20, 171. [Google Scholar] [CrossRef] [Green Version]
- Capecchi, M.R. The new mouse genetics: Altering the genome by gene targeting. Trends Genet. 1989, 5, 70–76. [Google Scholar] [CrossRef]
- Quadros, R.M.; Harms, D.W.; Ohtsuka, M.; Gurumurthy, C.B. Insertion of sequences at the original provirus integration site of mouse ROSA26 locus using the CRISPR/Cas9 system. FEBS Open Bio. 2015, 5, 191–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codner, G.F.; Mianne, J.; Caulder, A.; Loeffler, J.; Fell, R.; King, R.; Allan, A.J.; Mackenzie, M.; Pike, F.J.; McCabe, C.V.; et al. Application of long single-stranded DNA donors in genome editing: Generation and validation of mouse mutants. BMC Biol. 2018, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Hustedt, N.; Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell. Biol. 2016, 19, 1–9. [Google Scholar] [CrossRef]
- Yang, D.; Scavuzzo, M.A.; Chmielowiec, J.; Sharp, R.; Bajic, A.; Borowiak, M. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 2016, 6, 21264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 2014, 3, e04766. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, P. Mammalian zygotic genome activation. Semin. Cell Dev. Biol. 2018, 84, 118–126. [Google Scholar] [CrossRef]
- Ma, M.; Zhuang, F.; Hu, X.; Wang, B.; Wen, X.Z.; Ji, J.F.; Xi, J.J. Efficient generation of mice carrying homozygous double-floxP alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res. 2017, 27, 578–581. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Mackley, V.A.; Rao, A.; Chong, A.T.; Dewitt, M.A.; Corn, J.E.; Murthy, N. Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. Elife 2017, 6. [Google Scholar]
- Savic, N.; Ringnalda, F.C.; Lindsay, H.; Berk, C.; Bargsten, K.; Li, Y.; Neri, D.; Robinson, M.D.; Ciaudo, C.; Hall, J.; et al. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. Elife 2018, 7. [Google Scholar]
- Roche, P.J.R.; Gytz, H.; Hussain, F.; Cameron, C.J.F.; Paquette, D.; Blanchette, M.; Dostie, J.; Nagar, B.; Akavia, U.D. Double-Stranded Biotinylated Donor Enhances Homology-Directed Repair in Combination with Cas9 Monoavidin in Mammalian Cells. CRISPR J. 2018, 1, 414–430. [Google Scholar] [CrossRef]
- Yoon, Y.; Wang, D.; Tai, P.W.L.; Riley, J.; Gao, G.; Rivera-Perez, J.A. Streamlined ex vivo and in vivo genome editing in mouse embryos using recombinant adeno-associated viruses. Nat. Commun. 2018, 9, 412. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, N.; Mizutani, E.; Sato, H.; Kasai, M.; Ogawa, A.; Suchy, F.; Yamaguchi, T.; Nakauchi, H. Intra-embryo Gene Cassette Knockin by CRISPR/Cas9-Mediated Genome Editing with Adeno-Associated Viral Vector. iScience 2018, 9, 286–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modzelewski, A.J.; Chen, S.; Willis, B.J.; Lloyd, K.C.K.; Wood, J.A.; He, L. Efficient mouse genome engineering by CRISPR-EZ technology. Nat. Protoc. 2018, 13, 1253–1274. [Google Scholar] [CrossRef] [PubMed]
- Sawatsubashi, S.; Joko, Y.; Fukumoto, S.; Matsumoto, T.; Sugano, S.S. Development of versatile non-homologous end joining-based knock-in module for genome editing. Sci. Rep. 2018, 8, 593. [Google Scholar] [CrossRef]
- Lackner, D.H.; Carre, A.; Guzzardo, P.M.; Banning, C.; Mangena, R.; Henley, T.; Oberndorfer, S.; Gapp, B.V.; Nijman, S.M.B.; Brummelkamp, T.R.; et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat. Commun. 2015, 6, 10237. [Google Scholar] [CrossRef]
- Maresca, M.; Lin, V.G.; Guo, N.; Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): Custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013, 23, 539–546. [Google Scholar] [CrossRef] [Green Version]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Wang, X.; Hu, X.; Liu, Z.; Liu, J.; Zhou, H.; Shen, X.; Wei, Y.; Huang, Z.; Ying, W.; et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017, 27, 801–814. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.P.; Li, X.L.; Li, G.H.; Chen, W.; Arakaki, C.; Botimer, G.D.; Baylink, D.; Zhang, L.; Wen, W.; Fu, Y.W.; et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017, 18, 35. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.; Kweon, J.; Kim, H.S.; Kim, J.S. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 2014, 11, 705–706. [Google Scholar] [CrossRef]
- Gierut, J.J.; Jacks, T.E.; Haigis, K.M. Strategies to achieve conditional gene mutation in mice. Cold Spring Harb. Protoc. 2014, 2014, 339–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Du, Y.; Xie, W.; Zhang, F.; Forrest, D.; Liu, C. Generation of Conditional Knockout Mice by Sequential Insertion of Two loxP Sites In Cis Using CRISPR/Cas9 and Single-Stranded DNA Oligonucleotides. Methods Mol. Biol. 2019, 1874, 191–210. [Google Scholar] [PubMed]
- Takeo, T.; Nakagata, N. Superovulation using the combined administration of inhibin antiserum and equine chorionic gonadotropin increases the number of ovulated oocytes in C57BL/6 female mice. PLoS ONE 2015, 10, e0128330. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Sakuma, T.; Nishimichi, N.; Yokosaki, Y.; Yanaka, N.; Takeo, T.; Nakagata, N.; Yamamoto, T. Ultra-superovulation for the CRISPR-Cas9-mediated production of gene-knockout, single-amino-acid-substituted, and floxed mice. Biol. Open. 2016, 5, 1142–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, K.; Fukagawa, T.; Takisawa, H.; Kakimoto, T.; Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 2009, 6, 917–922. [Google Scholar] [CrossRef]
- Nabet, B.; Roberts, J.M.; Buckley, D.L.; Paulk, J.; Dastjerdi, S.; Yang, A.; Leggett, A.L.; Erb, M.A.; Lawlor, M.A.; Souza, A.; et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 2018, 14, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.J.; Macdonald, L.E.; Stevens, S.; Karow, M.; Dore, A.T.; Pobursky, K.; Huang, T.T.; Poueymirou, W.T.; Esau, L.; Meola, M.; et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc. Natl. Acad. Sci. USA 2014, 111, 5153–5158. [Google Scholar] [CrossRef] [Green Version]
- Leidy-Davis, T.; Cheng, K.; Goodwin, L.O.; Morgan, J.L.; Juan, W.C.; Roca, X.; Ong, S.T.; Bergstrom, D.E. Viable Mice with Extensive Gene Humanization (25-kbp) Created Using Embryonic Stem Cell/Blastocyst and CRISPR/Zygote Injection Approaches. Sci. Rep. 2018, 8, 15028. [Google Scholar] [CrossRef]
- Levine, M.S.; Cepeda, C.; Hickey, M.A.; Fleming, S.M.; Chesselet, M.F. Genetic mouse models of Huntington’s and Parkinson’s diseases: Illuminating but imperfect. Trends Neurosci. 2004, 27, 691–697. [Google Scholar] [CrossRef]
- Yan, S.; Tu, Z.; Liu, Z.; Fan, N.; Yang, H.; Yang, S.; Yang, W.; Zhao, Y.; Ouyang, Z.; Lai, C.; et al. A Huntingtin Knockin Pig Model Recapitulates Features of Selective Neurodegeneration in Huntington’s Disease. Cell 2018, 173, 989–1002.e13. [Google Scholar] [CrossRef] [Green Version]
- Lea, R.A.; Niakan, K.K. Human germline genome editing. Nat. Cell Biol. 2019, 21, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
| Technique | Repair Mechanism | Characterizing Features | Primary Benefits |
|---|---|---|---|
| Easi-CRISPR [52,53] | HDR | Long single-stranded DNA donor with short homology arms | Achieves large fragment knock-in with efficiencies similar to small ssODN-based knock-ins |
| 2C-HR-CRISPR [54] | HR | Microinjection into 2-cell staged embryo Template recruitment to the double-stranded break | Increased HR rates in 2-cell staged embryos lead to higher knock-in efficiency |
| CRISPR-READI [55] | HDR | Cas9 and gRNA electroporated into embryos followed by AAV-mediated delivery of donor template | Delivery of editing components does not require zygote microinjection |
| Tild-CRISPR [56] | HMEJ | Long, pre-linearized double-stranded donor with 800bp of homology | Linear double-stranded DNA donor overcomes size limitations of single-stranded DNA donors |
| CRIS-PITCh [57,58] | MMEJ | Plasmid-based donor with short homology arms linearized via Cas9 in vivo | Functions independently of HR, and requires minimal homology arm sequences |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erwood, S.; Gu, B. Embryo-Based Large Fragment Knock-in in Mammals: Why, How and What’s Next. Genes 2020, 11, 140. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11020140
Erwood S, Gu B. Embryo-Based Large Fragment Knock-in in Mammals: Why, How and What’s Next. Genes. 2020; 11(2):140. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11020140
Chicago/Turabian StyleErwood, Steven, and Bin Gu. 2020. "Embryo-Based Large Fragment Knock-in in Mammals: Why, How and What’s Next" Genes 11, no. 2: 140. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11020140




