Identification of Age-Specific and Common Key Regulatory Mechanisms Governing Eggshell Strength in Chicken Using Random Forests
Abstract
:1. Introduction
2. Materials and Methods
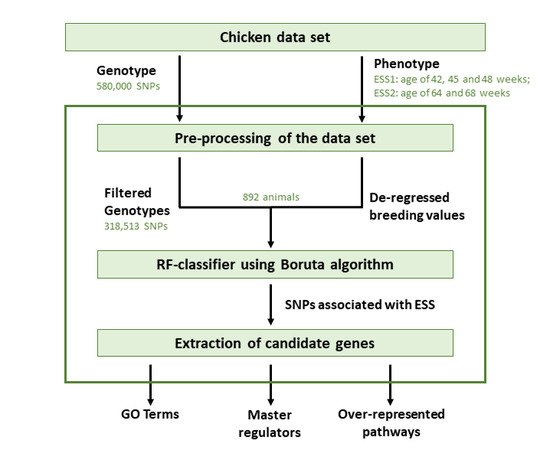
2.1. Chicken Dataset
2.2. Association Analysis Using Random Forests
2.3. Gene Set Analysis
2.4. Identification of Master Regulators and Over-Represented Pathways
3. Results
3.1. Gene Set Analysis
3.2. Identification of Master Regulators
3.3. Identification of Over-Represented Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Bain, M.; Nys, Y.; Dunn, I. Increasing persistency in lay and stabilising egg quality in longer laying cycles. What are the challenges? Br. Poult. Sci. 2016, 57, 330–338. [Google Scholar] [CrossRef] [Green Version]
- Pottgüter, R. Feeding laying hens to 100 weeks of age. Lohmann Inf. 2016, 50, 18–21. [Google Scholar]
- Chien, Y.C.; Hincke, M.; McKee, M. Ultrastructure of avian eggshell during resorption following egg fertilization. J. Struct. Biol. 2009, 168, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Nys, Y.; Bain, M.; Van Immerseel, F. Improving the Safety and Quality of Eggs and Egg Products: Volume 1: Egg Chemistry, Production and Consumption; Elsevier: Cambridge, UK, 2011. [Google Scholar]
- Brionne, A.; Nys, Y.; Hennequet-Antier, C.; Gautron, J. Hen uterine gene expression profiling during eggshell formation reveals putative proteins involved in the supply of minerals or in the shell mineralization process. BMC Genom. 2014, 15, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, K.; Maček, B.; Olsen, J.V. Proteomic analysis of the acid-soluble organic matrix of the chicken calcified eggshell layer. Proteomics 2006, 6, 3801–3810. [Google Scholar] [CrossRef]
- Mann, K.; Olsen, J.V.; Maček, B.; Gnad, F.; Mann, M. Phosphoproteins of the chicken eggshell calcified layer. Proteomics 2007, 7, 106–115. [Google Scholar] [CrossRef]
- Yin, Z.; Lian, L.; Zhu, F.; Zhang, Z.H.; Hincke, M.; Yang, N.; Hou, Z.C. The transcriptome landscapes of ovary and three oviduct segments during chicken (Gallus gallus) egg formation. Genomics 2020, 112, 243–251. [Google Scholar] [CrossRef]
- Crosara, F.S.G.; Pereira, V.J.; Lellis, C.G.; Barra, K.C.; Santos, S.K.A.D.; Souza, L.C.G.M.D.; Morais, T.A.D.; Litz, F.; Limão, V.A.; Braga, P.F.S.; et al. Is the Eggshell Quality Influenced by the Egg Weight or the Breeder Age? Braz. J. Poult. Sci. 2019, 21. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Wu, R. Mapping complex traits as a dynamic system. Phys. Life Rev. 2015, 13, 155–185. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, Z.; Wang, S.; Li, H. Progress of genome wide association study in domestic animals. J. Anim. Sci. Biotechnol. 2012, 3, 26. [Google Scholar] [CrossRef] [Green Version]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [Green Version]
- Schmid, M.; Bennewitz, J. Invited review: Genome-wide association analysis for quantitative traits in livestock–a selective review of statistical models and experimental designs. Arch. Fuer Tierz. 2017, 60, 335. [Google Scholar] [CrossRef]
- Johnson, R.C.; Nelson, G.W.; Troyer, J.L.; Lautenberger, J.A.; Kessing, B.D.; Winkler, C.A.; O’Brien, S.J. Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC Genom. 2010, 11, 724. [Google Scholar] [CrossRef] [Green Version]
- Bush, W.S.; Moore, J.H. Genome-wide association studies. PLoS Comput. Biol. 2012, 8, e1002822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, D.; Fan, C.C.; Frei, O.; Shadrin, A.A.; Smeland, O.B.; Sundar, V.; Andreassen, O.A.; Dale, A.M. Estimating inflation in GWAS summary statistics due to variance distortion from cryptic relatedness. BioRxiv 2017, 164939. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, A.I. Solving the missing heritability problem. PLoS Genet. 2019, 15, e1008222. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chen, F.; Zhai, R.; Lin, X.; Wang, Z.; Su, L.; Christiani, D.C. Correction for population stratification in random forest analysis. Int. J. Epidemiol. 2012, 41, 1798–1806. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.T.; Huang, J.Z.; Wu, Q.; Nguyen, T.T.; Li, M.J. Genome-wide association data classification and SNPs selection using two-stage quality-based Random Forests. BMC Genom. 2015, 16, S5. [Google Scholar] [CrossRef] [Green Version]
- Armero, C.; Cabras, S.; Castellanos, M.E.; Quirós, A. Two-Stage Bayesian Approach for GWAS With Known Genealogy. J. Comput. Graph. Stat. 2019, 28, 197–204. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, W.; Bellis, E.S.; Stubblefield, J.; Causey, J.; Qualls, J.; Walker, K. Minor QTLs mining through the combination of GWAS and machine learning feature selection. BioRxiv 2019, 712190. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, D.; He, F.; Wang, J.; Joshi, T.; Xu, D. Phenotype prediction and genome-wide association study using deep convolutional neural network of soybean. Front. Genet. 2019, 10, 1091. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Schwender, H.; Keith, J.; Nunkesser, R.; Mengersen, K.; Macrossan, P. Methods for identifying SNP interactions: A review on variations of Logic Regression, Random Forest and Bayesian logistic regression. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011, 8, 1580–1591. [Google Scholar] [CrossRef] [PubMed]
- Van der Heide, E.; Veerkamp, R.; van Pelt, M.; Kamphuis, C.; Athanasiadis, I.; Ducro, B. Comparing regression, naive Bayes, and random forest methods in the prediction of individual survival to second lactation in Holstein cattle. J. Dairy Sci. 2019, 102, 9409–9421. [Google Scholar] [CrossRef] [PubMed]
- Brieuc, M.S.; Waters, C.D.; Drinan, D.P.; Naish, K.A. A practical introduction to Random Forest for genetic association studies in ecology and evolution. Mol. Ecol. Resour. 2018, 18, 755–766. [Google Scholar] [CrossRef]
- Nguyen, T.; Le, L. Detection of SNP-SNP Interactions in Genome-wide Association Data Using Random Forests and Association Rules. In Proceedings of the 2018 12th International Conference on Software, Knowledge, Information Management & Applications (SKIMA), Phnom Penh, Cambodia, 3–5 December 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–7. [Google Scholar]
- Romagnoni, A.; Jégou, S.; Van Steen, K.; Wainrib, G.; Hugot, J.P. Comparative performances of machine learning methods for classifying Crohn Disease patients using genome-wide genotyping data. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef]
- Erbe, M.; Cavero, D.; Weigend, A.; Weigend, S.; Pausch, H.; Preisinger, R.; Simianer, H. Genomic prediction in laying hens. In Proceedings of the 8th European Symposium on Poultry Genetics, Venice, Italy, 25–27 September 2013. [Google Scholar]
- Ni, G.; Strom, T.M.; Pausch, H.; Reimer, C.; Preisinger, R.; Simianer, H.; Erbe, M. Comparison among three variant callers and assessment of the accuracy of imputation from SNP array data to whole-genome sequence level in chicken. BMC Genom. 2015, 16, 824. [Google Scholar] [CrossRef] [Green Version]
- Ni, G.; Cavero, D.; Fangmann, A.; Erbe, M.; Simianer, H. Whole-genome sequence-based genomic prediction in laying chickens with different genomic relationship matrices to account for genetic architecture. Genet. Sel. Evol. 2017, 49, 8. [Google Scholar] [CrossRef] [Green Version]
- Garrick, D.J.; Taylor, J.F.; Fernando, R.L. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 2009, 41, 55. [Google Scholar] [CrossRef] [Green Version]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the Boruta package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kursa, M.B.; Jankowski, A.; Rudnicki, W.R. Boruta–a system for feature selection. Fundam. Informaticae 2010, 101, 271–285. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. The all relevant feature selection using random forest. arXiv 2011, arXiv:1106.5112. [Google Scholar]
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011. [Google Scholar] [CrossRef]
- Ayalew, Y.; Gültas, M.; Effa, K.; Hanotte, O.H.; Schmitt, A. Identification of candidate signature genes and key regulators associated with Trypanotolerance in the Sheko Breed. Front. Genet. 2019, 10, 1095. [Google Scholar]
- Wlochowitz, D.; Haubrock, M.; Arackal, J.; Bleckmann, A.; Wolff, A.; Beißbarth, T.; Wingender, E.; Gültas, M. Computational identification of key regulators in two different colorectal cancer cell lines. Front. Genet. 2016, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Wingender, E.; Kel, A. geneXplain—eine integrierte Bioinformatik-Plattform. BIOspektrum 2012, 18, 554–556. [Google Scholar] [CrossRef]
- Koschmann, J.; Bhar, A.; Stegmaier, P.; Kel, A.; Wingender, E. “Upstream analysis”: An integrated promoter-pathway analysis approach to causal interpretation of microarray data. Microarrays 2015, 4, 270–286. [Google Scholar] [CrossRef]
- Rzhetsky, A.; Iossifov, I.; Koike, T.; Krauthammer, M.; Kra, P.; Morris, M.; Yu, H.; Duboué, P.A.; Weng, W.; Wilbur, W.J.; et al. GeneWays: A system for extracting, analyzing, visualizing, and integrating molecular pathway data. J. Biomed. Inform. 2004, 37, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Krull, M.; Pistor, S.; Voss, N.; Kel, A.; Reuter, I.; Kronenberg, D.; Michael, H.; Schwarzer, K.; Potapov, A.; Choi, C.; et al. TRANSPATH®: An information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res. 2006, 34, D546–D551. [Google Scholar] [CrossRef] [Green Version]
- Jonchère, V.; Brionne, A.; Gautron, J.; Nys, Y. Identification of uterine ion transporters for mineralisation precursors of the avian eggshell. BMC Physiol. 2012, 12, 10–51. [Google Scholar] [CrossRef] [Green Version]
- Chakraborti, S.; Dhalla, N.S. Regulation of Membrane Na+-K+ ATPase; Springer: Heidelberg, Germany, 2016. [Google Scholar]
- Colbran, R.J. Targeting of calcium/calmodulin-dependent protein kinase II. Biochem. J. 2004, 378, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.B.; Watanuki, M.; Kim, S.; Shevde, N.K.; Pike, J.W. The human transient receptor potential vanilloid type 6 distal promoter contains multiple vitamin D receptor binding sites that mediate activation by 1, 25-dihydroxyvitamin D3 in intestinal cells. Mol. Endocrinol. 2006, 20, 1447–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogata, K.; Jeong, S.Y.; Murakami, H.; Hashida, H.; Suzuki, T.; Masuda, N.; Hirai, M.; Isahara, K.; Uchiyama, Y.; Goto, J.; et al. Cloning and expression study of the mouse tetrodotoxin-resistant voltage-gated sodium channel α subunit NaT/Scn11a. Biochem. Biophys. Res. Commun. 2000, 267, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Seda, M.; Pinto, F.M.; Wray, S.; Cintado, C.G.; Noheda, P.; Buschmann, H.; Candenas, L. Functional and molecular characterization of voltage-gated sodium channels in uteri from nonpregnant rats. Biol. Reprod. 2007, 77, 855–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.F.; Hou, Z.C.; Yi, G.Q.; Xu, G.Y.; Yang, N. The sodium channel gene family is specifically expressed in hen uterus and associated with eggshell quality traits. BMC Genet. 2013, 14, 90. [Google Scholar] [CrossRef] [Green Version]
- Koepsell, H. The SLC22 family with transporters of organic cations, anions and zwitterions. Mol. Asp. Med. 2013, 34, 413–435. [Google Scholar] [CrossRef]
- Chowdhury, S.; Smith, T. Dietary interaction of 1, 4-diaminobutane (putrescine) and calcium on eggshell quality and performance in laying hens. Poult. Sci. 2002, 81, 84–91. [Google Scholar] [CrossRef]
- Shinki, T.; Tanaka, H.; Takito, J.; Yamaguchi, A.; Nakamura, Y.; Yoshiki, S.; Suda, T. Putrescine is involved in the vitamin D action in chick intestine. Gastroenterology 1991, 100, 113–122. [Google Scholar] [CrossRef]
- Altimimi, H.F.; Schnetkamp, P.P. Na+/Ca2+-K+ exchangers (NCKX): Functional properties and physiological roles. Channels 2007, 1, 62–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadi, J.; Jung, S.H.; Lee, M.J.; Jami, A.; Ruthala, K.; Kim, K.M.; Cho, N.H.; Jung, H.S.; Kim, C.H.; Lim, S.K. The transcription factor protein Sox11 enhances early osteoblast differentiation by facilitating proliferation and the survival of mesenchymal and osteoblast progenitors. J. Biol. Chem. 2013, 288, 25400–25413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ELAROUSSI, M.A.; FORTE, L.R.; EBER, S.L.; BIELLIER, H.V. Calcium Homeostasis in the Laying Hen.: 1. Age and Dietary Calcium Effects. Poult. Sci. 1994, 73, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Håkelien, A.M.; Bryne, J.C.; Harstad, K.G.; Lorenz, S.; Paulsen, J.; Sun, J.; Mikkelsen, T.S.; Myklebost, O.; Meza-Zepeda, L.A. The regulatory landscape of osteogenic differentiation. Stem Cells 2014, 32, 2780–2793. [Google Scholar] [CrossRef] [PubMed]
- Scheidegger, E.P.; Sternberg, L.R.; Roth, J.; Lowe, J.B. A human STX cDNA confers polysialic acid expression in mammalian cells. J. Biol. Chem. 1995, 270, 22685–22688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, T.; Munakata, K.; Adachi, S.; Hatta, H.; Nakamura, T.; Kato, T.; Kim, M. Chalaza and egg yolk membrane as excellent sources of sialic acid (N-acetylneuraminic acid) for an industrial-scale preparation. Jpn. J. Zootech. Sci. 1990, 61, 277–282. [Google Scholar]
- Nakano, K.; Nakano, T.; Ahn, D.; Sim, J. Sialic acid contents in chicken eggs and tissues. Can. J. Anim. Sci. 1994, 74, 601–606. [Google Scholar] [CrossRef]
- Nakano, T.; Ikawa, N.; Ozimek, L. Chemical composition of chicken eggshell and shell membranes. Poult. Sci. 2003, 82, 510–514. [Google Scholar] [CrossRef]
- Du, J.; Hincke, M.T.; Rose-Martel, M.; Hennequet-Antier, C.; Brionne, A.; Cogburn, L.A.; Nys, Y.; Gautron, J. Identifying specific proteins involved in eggshell membrane formation using gene expression analysis and bioinformatics. BMC Genom. 2015, 16, 792. [Google Scholar] [CrossRef] [Green Version]
- Jonchère, V.; Réhault-Godbert, S.; Hennequet-Antier, C.; Cabau, C.; Sibut, V.; Cogburn, L.A.; Nys, Y.; Gautron, J. Gene expression profiling to identify eggshell proteins involved in physical defense of the chicken egg. BMC Genom. 2010, 11, 57. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, T.A.; Suso, H.P.; Hincke, M.T. Experimental datasets on processed eggshell membrane powder for wound healing. Data Brief 2019, 26, 104457. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.A.; Suso, H.P.; Hincke, M.T. In-depth comparative analysis of the chicken eggshell membrane proteome. J. Proteom. 2017, 155, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.m.; Kim, W.Y.; Nam, S.A.; Choi, A.R.; Kim, H.; Kim, Y.K.; Kim, H.S.; Kim, J. Role of PROX1 in the transforming ascending thin limb of Henle’s loop during mouse kidney development. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [Green Version]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogasawara, T.; Mori, Y.; Abe, M.; Suenaga, H.; Kawase-Koga, Y.; Saijo, H.; Takato, T. Role of cyclin-dependent kinase (Cdk) 6 in osteoblast, osteoclast, and chondrocyte differentiation and its potential as a target of bone regenerative medicine. Oral Sci. Int. 2011, 8, 2–6. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, C. Overview of bone biology in the egg-laying hen. Poult. Sci. 2004, 83, 193–199. [Google Scholar] [CrossRef]
- Bar, A. Calcium transport in strongly calcifying laying birds: Mechanisms and regulation. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009, 152, 447–469. [Google Scholar] [CrossRef]
- Ono, K.; Han, J. The p38 signal transduction pathway activation and function. Cell. Signal. 2000, 12, 1–13. [Google Scholar] [CrossRef]
- Suzanne, M.; Irie, K.; Glise, B.; Agnès, F.; Mori, E.; Matsumoto, K.; Noselli, S. The Drosophila p38 MAPK pathway is required during oogenesis for egg asymmetric development. Genes Dev. 1999, 13, 1464–1474. [Google Scholar] [CrossRef] [Green Version]
- Zelcer, N.; Tontonoz, P. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Investig. 2006, 116, 607–614. [Google Scholar] [CrossRef] [Green Version]
- Vaya, J.; Schipper, H.M. Oxysterols, cholesterol homeostasis, and Alzheimer disease. J. Neurochem. 2007, 102, 1727–1737. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, W.J.; Abdel-Khalik, J.; Crick, P.J.; Yutuc, E.; Wang, Y. New methods for analysis of oxysterols and related compounds by LC–MS. J. Steroid Biochem. Mol. Biol. 2016, 162, 4–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, M.; Hansen, K. Role of estrogen in avian osteoporosis. Poult. Sci. 2004, 83, 200–206. [Google Scholar] [CrossRef]
- Mackrill, J.J. Oxysterols and calcium signal transduction. Chem. Phys. Lipids 2011, 164, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Kha, H.T.; Basseri, B.; Shouhed, D.; Richardson, J.; Tetradis, S.; Hahn, T.J.; Parhami, F. Oxysterols regulate differentiation of mesenchymal stem cells: Pro-bone and anti-fat. J. Bone Miner. Res. 2004, 19, 830–840. [Google Scholar] [CrossRef] [Green Version]
- Frederick, J.P.; Liberati, N.T.; Waddell, D.S.; Shi, Y.; Wang, X.F. Transforming growth factor β-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol. Cell. Biol. 2004, 24, 2546–2559. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.R.; Kang, Y.; Siegel, P.M.; Massagué, J. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 2002, 110, 19–32. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.Y.; Alliston, T. Regulation of postnatal bone homeostasis by TGFβ. BoneKEy Rep. 2013, 2. [Google Scholar] [CrossRef] [Green Version]
- Bell, D.M.; Leung, K.K.; Wheatley, S.C.; Ng, L.J.; Zhou, S.; Ling, K.W.; Sham, M.H.; Koopman, P.; Tam, P.P.; Cheah, K.S. SOX9 directly regulates the type-ll collagen gene. Nat. Genet. 1997, 16, 174–178. [Google Scholar] [CrossRef]
- Massagué, J.; Chen, Y.G. Controlling TGF-β signaling. Genes Dev. 2000, 14, 627–644. [Google Scholar]
- Lönn, P.; Vanlandewijck, M.; Raja, E.; Kowanetz, M.; Watanabe, Y.; Kowanetz, K.; Vasilaki, E.; Heldin, C.H.; Moustakas, A. Transcriptional induction of salt-inducible kinase 1 by transforming growth factor β leads to negative regulation of type I receptor signaling in cooperation with the Smurf2 ubiquitin ligase. J. Biol. Chem. 2012, 287, 12867–12878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stow, L.R.; Jacobs, M.E.; Wingo, C.S.; Cain, B.D. Endothelin-1 gene regulation. FASEB J. 2011, 25, 16–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strait, K.A.; Stricklett, P.K.; Kohan, J.L.; Miller, M.B.; Kohan, D.E. Calcium regulation of endothelin-1 synthesis in rat inner medullary collecting duct. Am. J. Physiol.-Ren. Physiol. 2007, 293, F601–F606. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.S.; Jung, K.I.; Choi, Y.L.; Choi, E.Y.; Lee, I.S.; Park, S.H.; Kim, T.J. Engagement of CD99 triggers the exocytic transport of ganglioside GM1 and the reorganization of actin cytoskeleton. FEBS Lett. 2003, 540, 217–222. [Google Scholar] [CrossRef] [Green Version]
- Pasello, M.; Manara, M.C.; Scotlandi, K. CD99 at the crossroads of physiology and pathology. J. Cell Commun. Signal. 2018, 12, 55–68. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Bellon, M.; Shelton, S.N.; Nicot, C. Tumor suppressors p53, p63TAα, p63TAy, p73α, and p73β use distinct pathways to repress telomerase expression. J. Biol. Chem. 2012, 287, 20737–20747. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.S.; Yun, J.; Chae, H.D.; Kim, J.M.; Kim, S.C.; Choi, T.S.; Shin, D.Y. p53 and its homologues, p63 and p73, induce a replicative senescence through inactivation of NF-Y transcription factor. Oncogene 2001, 20, 5818–5825. [Google Scholar] [CrossRef] [Green Version]
- Alimov, A.P.; Park-Sarge, O.K.; Sarge, K.D.; Malluche, H.H.; Koszewski, N.J. Transactivation of the parathyroid hormone promoter by specificity proteins and the nuclear factor Y complex. Endocrinology 2005, 146, 3409–3416. [Google Scholar] [CrossRef] [Green Version]
- Jääskeläinen, T.; Huhtakangas, J.; Mäenpää, P. Negative regulation of human parathyroid hormone gene promoter by vitamin D3 through nuclear factor Y. Biochem. Biophys. Res. Commun. 2005, 328, 831–837. [Google Scholar] [CrossRef]
- Josephs, E.B.; Stinchcombe, J.R.; Wright, S.I. What can genome-wide association studies tell us about the evolutionary forces maintaining genetic variation for quantitative traits? New Phytol. 2017, 214, 21–33. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Zhang, N.; Wang, Y.G.; George, A.W.; Reverter, A.; Li, Y. Genomic prediction of breeding values using a subset of SNPs identified by three machine learning methods. Front. Genet. 2018, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Mikšík, I.; Eckhardt, A.; Sedláková, P.; Mikulikova, K. Proteins of insoluble matrix of avian (Gallus gallus) eggshell. Connect. Tissue Res. 2007, 48, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mikšík, I.; Sedláková, P.; Lacinová, K.; Pataridis, S.; Eckhardt, A. Determination of insoluble avian eggshell matrix proteins. Anal. Bioanal. Chem. 2010, 397, 205–214. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Sillanpää, M.J.; Silventoinen, K.; Kaprio, J.; Pitkäniemi, J. Estimating modifying effect of age on genetic and environmental variance components in twin models. Genetics 2016, 202, 1313–1328. [Google Scholar] [CrossRef] [Green Version]
- Elks, C.E.; Den Hoed, M.; Zhao, J.H.; Sharp, S.J.; Wareham, N.J.; Loos, R.J.; Ong, K.K. Variability in the heritability of body mass index: A systematic review and meta-regression. Front. Endocrinol. 2012, 3, 29. [Google Scholar] [CrossRef] [Green Version]
- Park, J.A.; Sohn, S.H. The Influence of Hen Aging on Eggshell Ultrastructure and Shell Mineral Components. Korean J. Food Sci. Anim. Resour. 2018, 38, 1080. [Google Scholar] [CrossRef] [Green Version]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g: Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- Neupane, M.; Geary, T.W.; Kiser, J.N.; Burns, G.W.; Hansen, P.J.; Spencer, T.E.; Neibergs, H.L. Loci and pathways associated with uterine capacity for pregnancy and fertility in beef cattle. PLoS ONE 2017, 12, e0188997. [Google Scholar] [CrossRef] [Green Version]
- Woldesemayat, A.A.; Ntwasa, M. Pathways and Network Based Analysis of Candidate Genes to Reveal Cross-Talk and Specificity in the Sorghum (Sorghum bicolor (L.) Moench) Responses to Drought and It’s Co-occurring Stresses. Front. Genet. 2018, 9, 557. [Google Scholar] [CrossRef] [Green Version]
- Ramayo-Caldas, Y.; Renand, G.; Ballester, M.; Saintilan, R.; Rocha, D. Multi-breed and multi-trait co-association analysis of meat tenderness and other meat quality traits in three French beef cattle breeds. Genet. Sel. Evol. 2016, 48, 37. [Google Scholar] [CrossRef] [Green Version]
- Kadarmideen, H.N.; von Rohr, P.; Janss, L.L. From genetical genomics to systems genetics: Potential applications in quantitative genomics and animal breeding. Mamm. Genome 2006, 17, 548–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| GO Term | GO Title | Number of Genes | Adjusted p-Value |
|---|---|---|---|
| GO:0005515 | protein binding | 281 | |
| GO:0005488 | binding | 331 | |
| GO:0043167 | ion binding | 155 | |
| GO:0000146 | microfilament motor activity | 5 | |
| GO:0003779 | actin binding | 20 | |
| GO:0032559 | adenyl ribonucleotide binding | 49 | |
| GO:0030554 | adenyl nucleotide binding | 49 | |
| GO:0044877 | macromolecular complex binding | 50 | |
| GO:0004683 | calmodulin-dependent protein kinase activity | 5 | |
| GO:0005524 | ATP binding | 47 | |
| GO:0042623 | ATPase activity, coupled | 16 | |
| GO:0008092 | cytoskeletal protein binding | 30 | |
| GO:0043168 | anion binding | 74 | |
| GO:0046983 | protein dimerization activity | 40 | |
| GO:0017016 | Ras GTPase binding | 12 |
| GO Term | GO Title | Number of Genes | Adjusted p-Value |
|---|---|---|---|
| GO:0005515 | protein binding | 168 | |
| GO:0022843 | voltage-gated cation channel activity | 9 | |
| GO:0005242 | inward rectifier potassium channel activity | 4 | |
| GO:0032549 | ribonucleoside binding | 40 | |
| GO:0000166 | nucleotide binding | 48 | |
| GO:0005524 | ATP binding | 34 | |
| GO:0001883 | purine nucleoside binding | 39 | |
| GO:0032559 | adenyl ribonucleotide binding | 34 | |
| GO:0005488 | binding | 199 | |
| GO:0030554 | adenyl nucleotide binding | 34 | |
| GO:0051427 | hormone receptor binding | 9 | |
| GO:0015276 | ligand-gated ion channel activity | 8 | |
| GO:0017076 | purine nucleotide binding | 39 | |
| GO:0022836 | gated channel activity | 12 | |
| GO:0036094 | small molecule binding | 50 |
| Pathway Name | Adjusted p-Value for ESS1 / ESS2 | Over-Represented in |
|---|---|---|
| E2F —/ Smad4 | / | ESS1, ESS2 |
| Endothelin-1 gene regulation | / - | ESS1 |
| G2/M phase (cyclin A:Cdk1) | / | ESS1, ESS2 |
| SMAD7, SIK1 gene induction | / - | ESS1 |
| oxysterol —>apoE | / | ESS1, ESS2 |
| LXR network | / | ESS1, ESS2 |
| p73alpha —/ NF-Y | - / | ESS2 |
| Sox9 —Smad3—>COL2A1 | / - | ESS1 |
| G1 phase (Cdk6) | / | ESS1, ESS2 |
| G1 phase (Cdk4) | / | ESS1, ESS2 |
| p38 pathway | / | ESS1, ESS2 |
| MIC2 signaling | - / | ESS2 |
| TGFbeta pathway | / - | ESS1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramzan, F.; Klees, S.; Schmitt, A.O.; Cavero, D.; Gültas, M. Identification of Age-Specific and Common Key Regulatory Mechanisms Governing Eggshell Strength in Chicken Using Random Forests. Genes 2020, 11, 464. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11040464
Ramzan F, Klees S, Schmitt AO, Cavero D, Gültas M. Identification of Age-Specific and Common Key Regulatory Mechanisms Governing Eggshell Strength in Chicken Using Random Forests. Genes. 2020; 11(4):464. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11040464
Chicago/Turabian StyleRamzan, Faisal, Selina Klees, Armin Otto Schmitt, David Cavero, and Mehmet Gültas. 2020. "Identification of Age-Specific and Common Key Regulatory Mechanisms Governing Eggshell Strength in Chicken Using Random Forests" Genes 11, no. 4: 464. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11040464