PlantMirP-Rice: An Efficient Program for Rice Pre-miRNA Prediction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Preparation
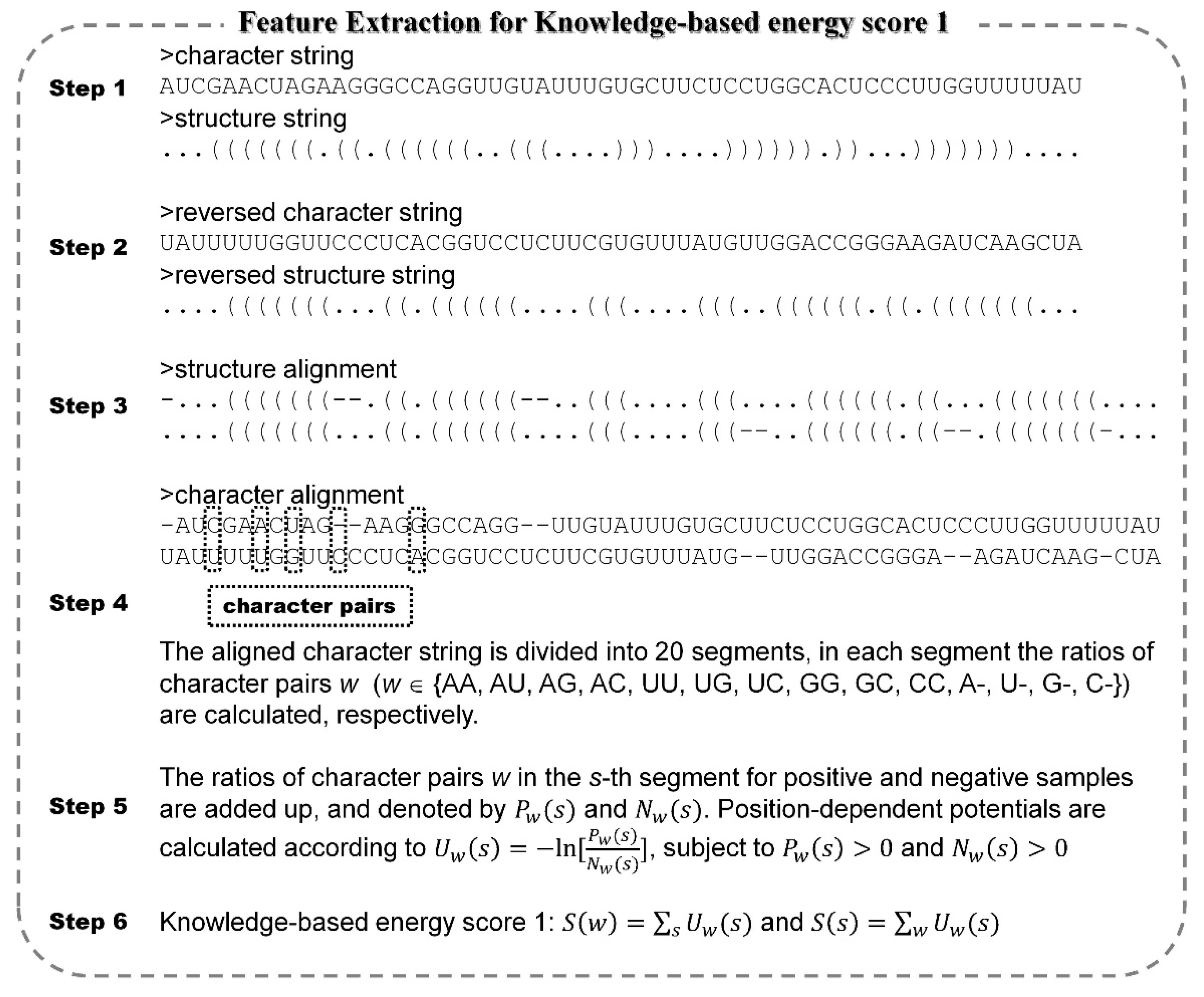
2.2. Feature Extraction
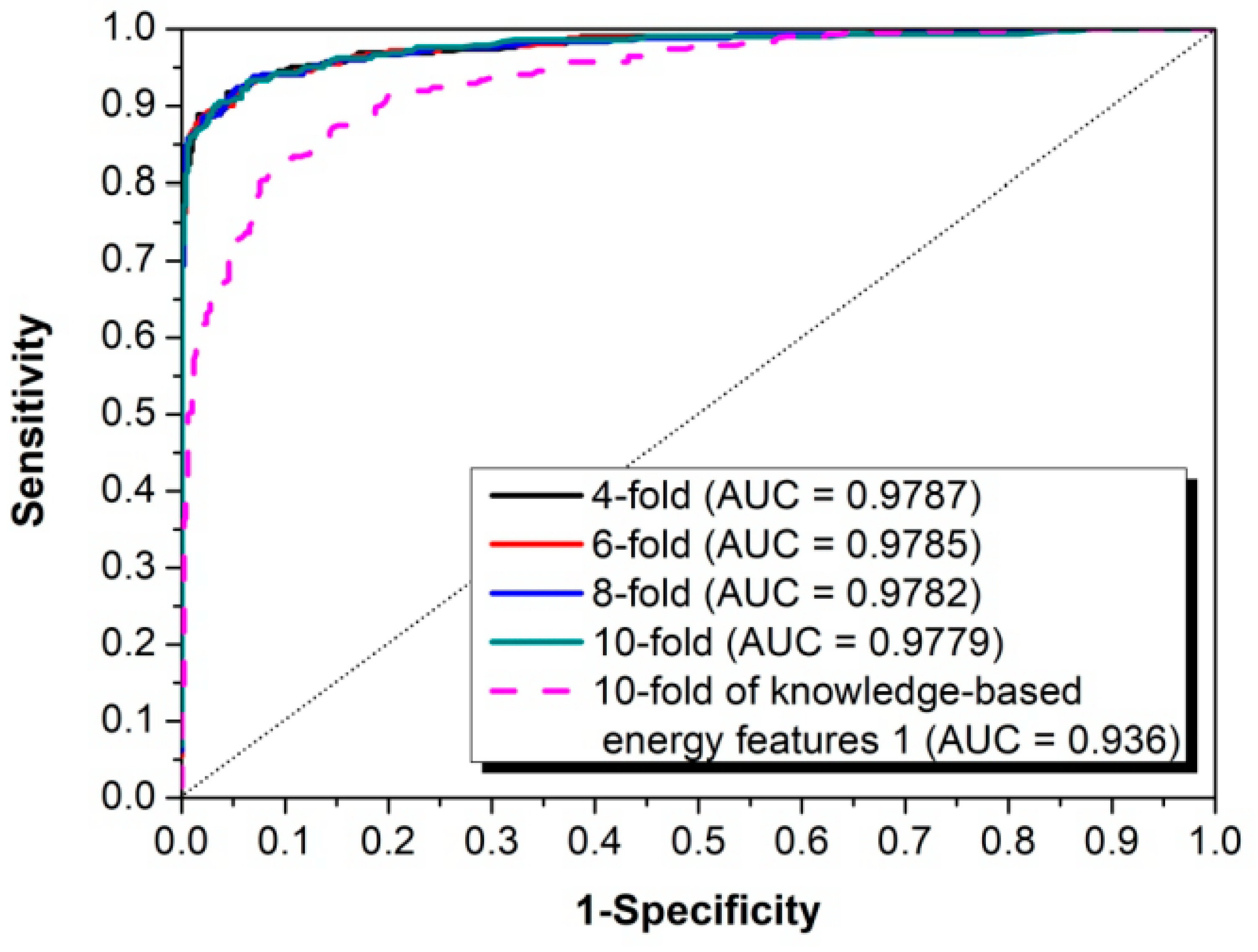
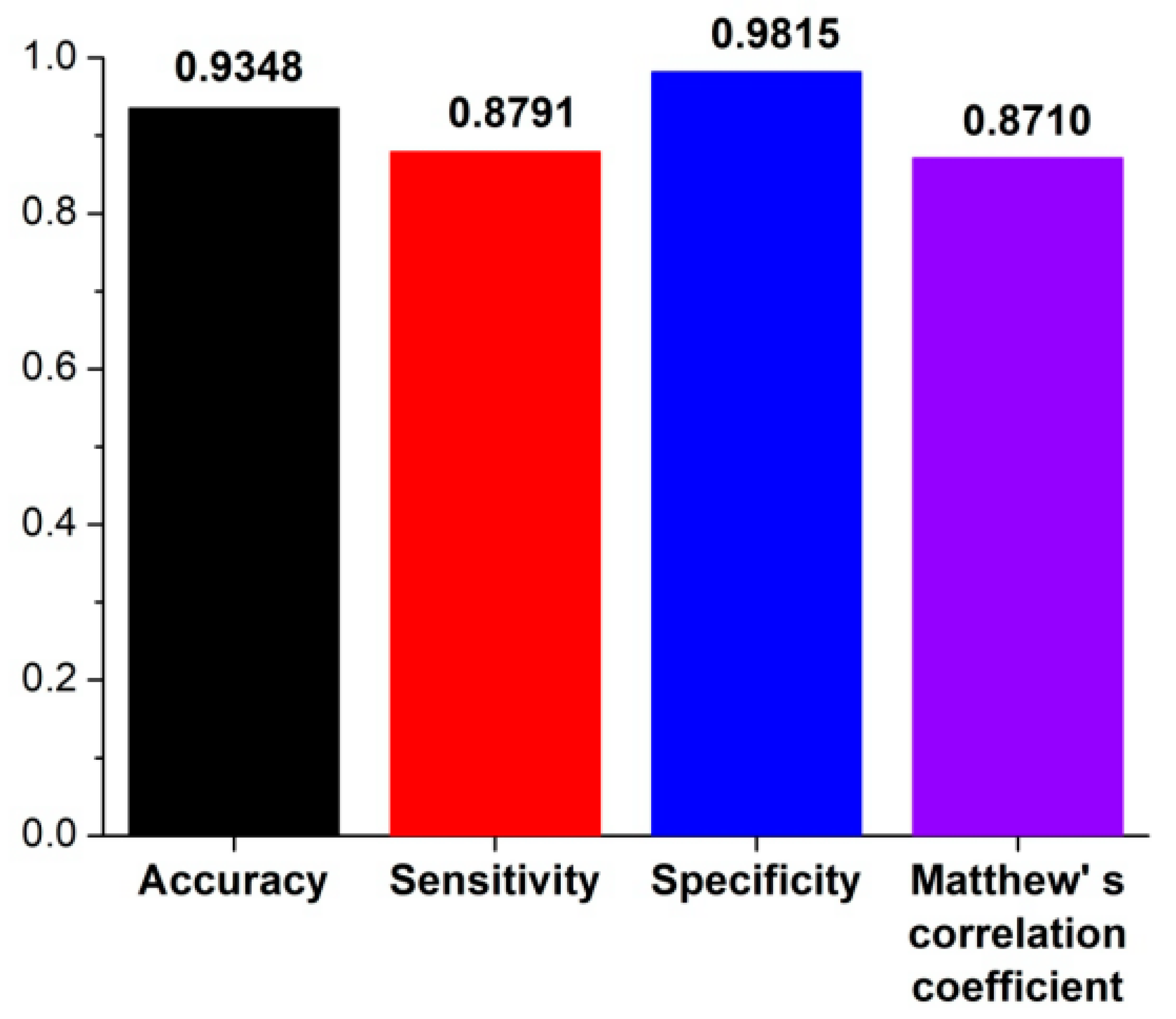
2.3. Performance Evaluation
2.4. Input, Output, Dependencies, Platforms, and Application Scenarios
3. Results
3.1. The Algorithm for the Prediction of Rice Pre-miRNAs
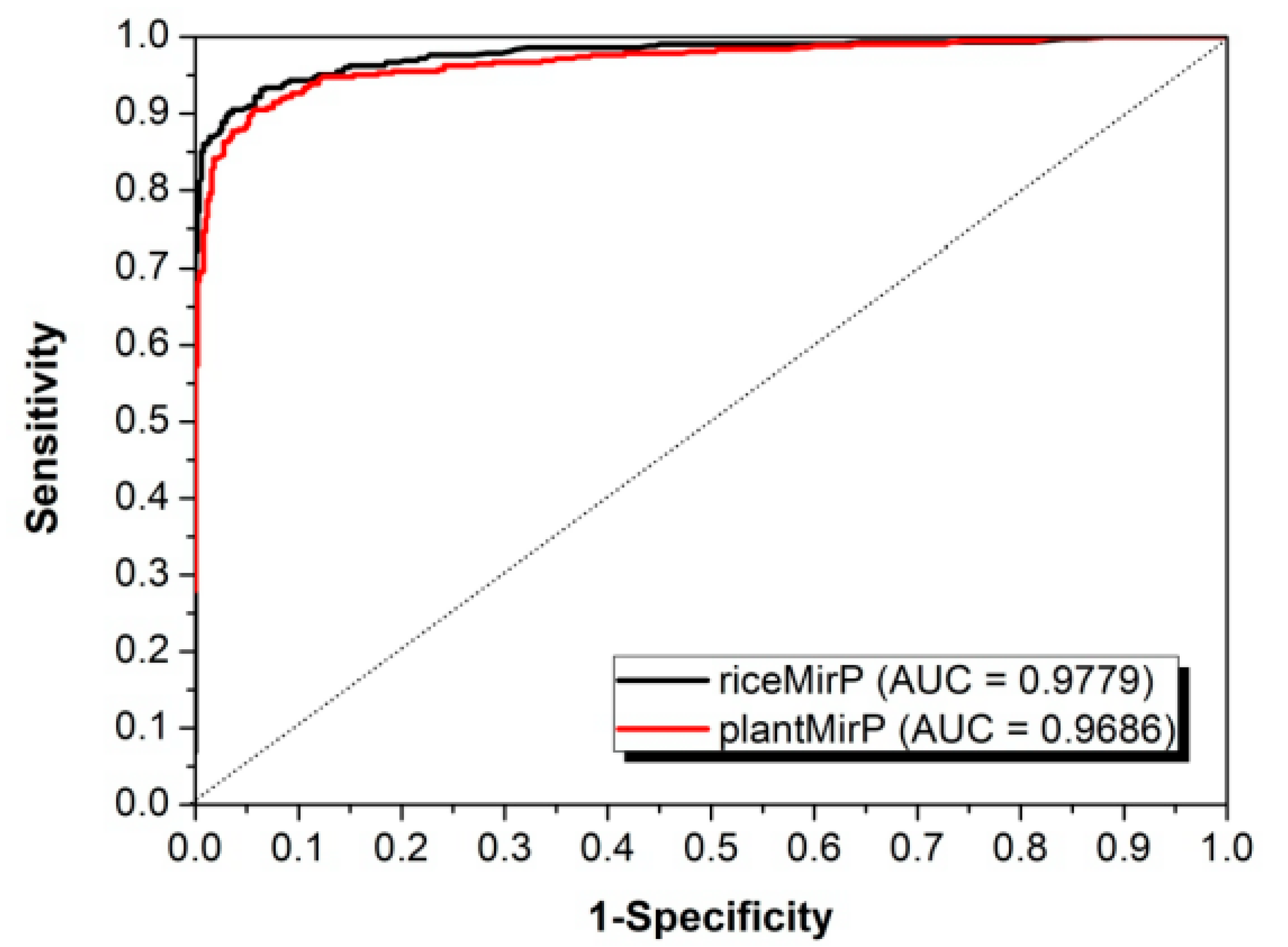
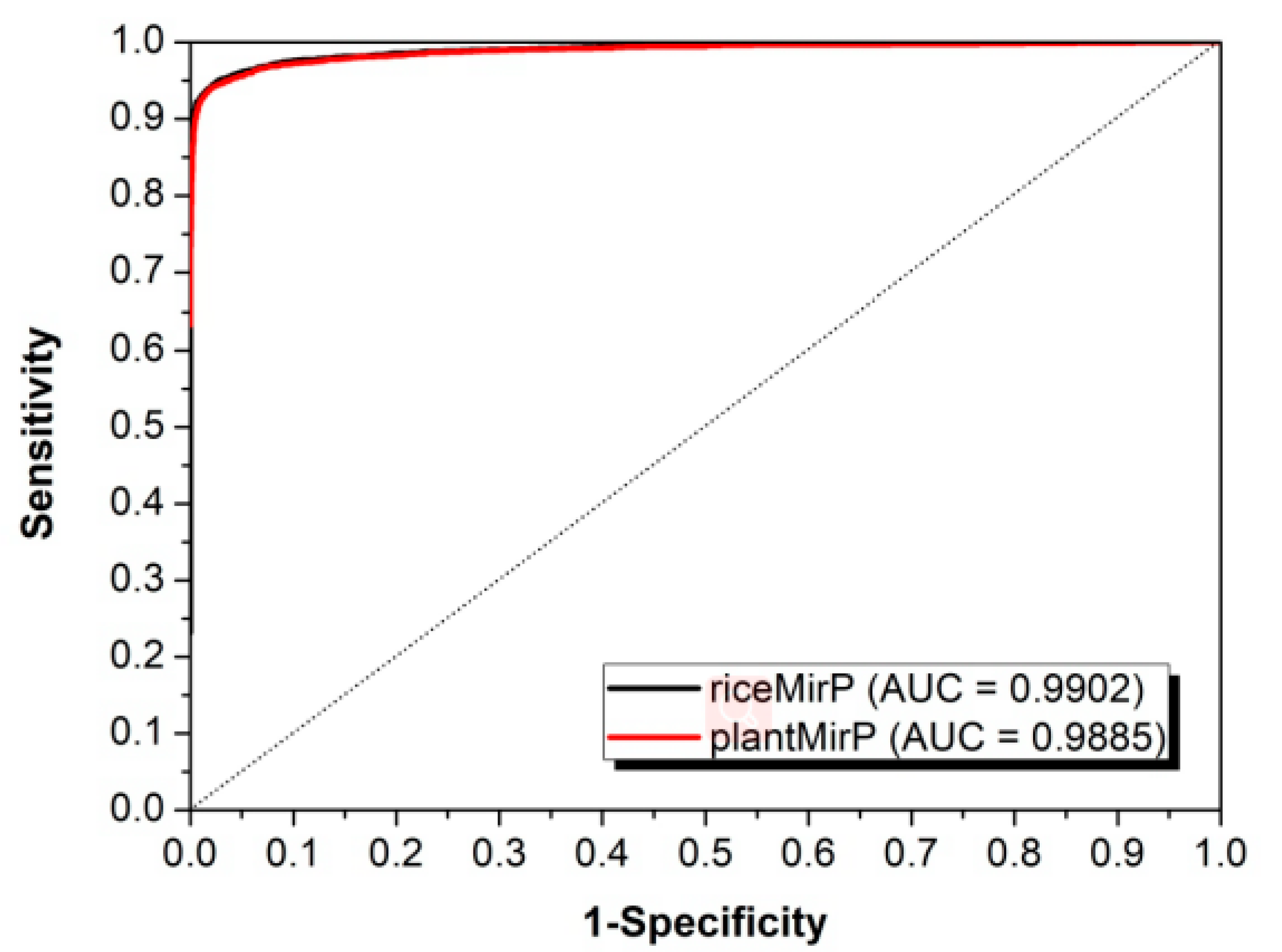
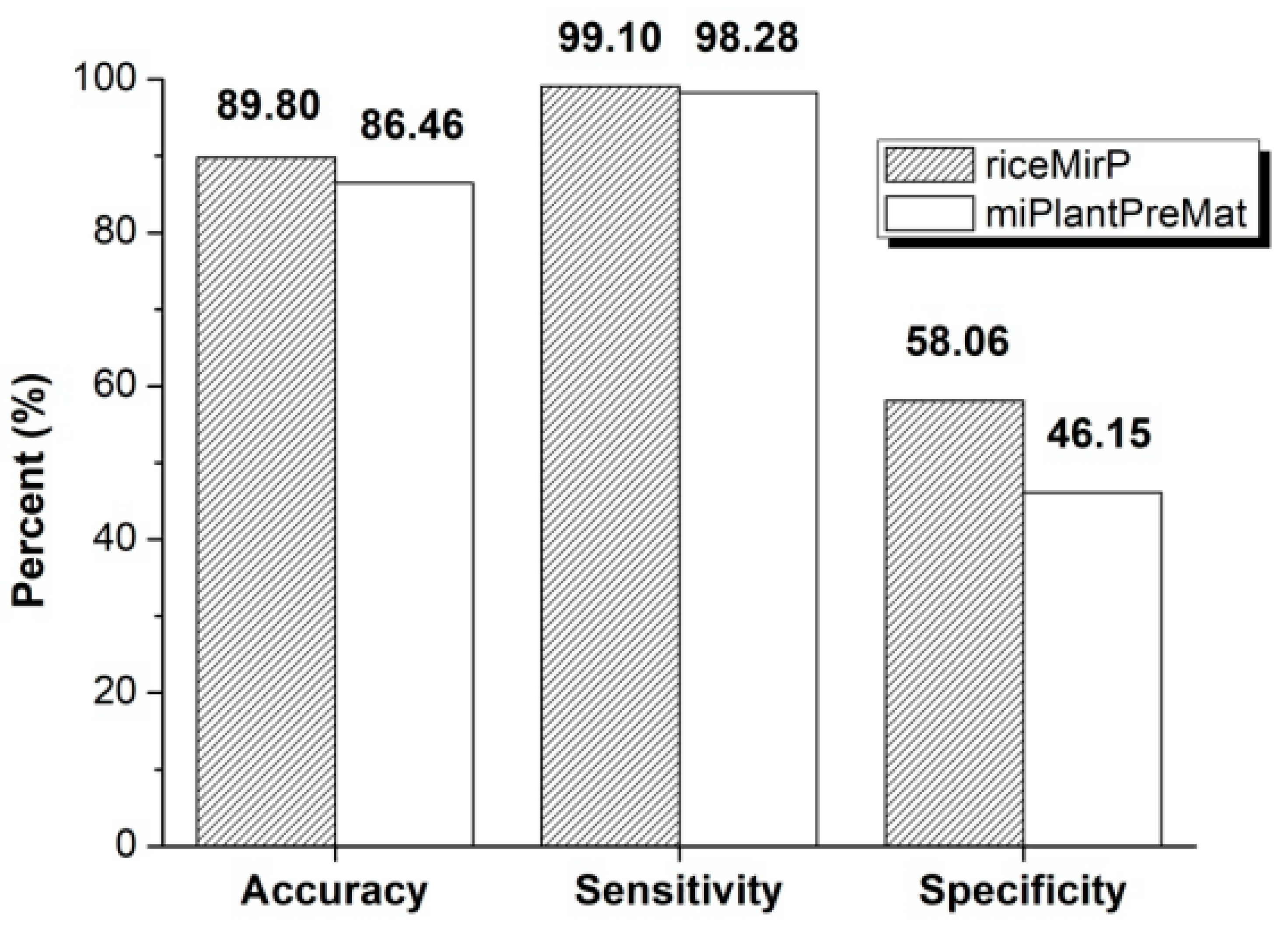
3.2. Comparison with Other Competitive Methods
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Teune, J.-H.; Steger, G. NOVOMIR: De Novo Prediction of MicroRNA-Coding Regions in a Single Plant-Genome. J. Nucleic Acids 2010, 2010, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Tao, Y.; Zhu, C. Emerging roles of microRNAs in the mediation of drought stress response in plants. J. Exp. Bot. 2013, 64, 3077–3086. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, J.; Liu, D.; Sun, C.; Luan, Y.-S. Prediction of plant pre-microRNAs and their microRNAs in genome-scale sequences using structure-sequence features and support vector machine. BMC Bioinform. 2014, 15, 423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D.G. A Plant miRNA Contributes to Antibacterial Resistance by Repressing Auxin Signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.; Feng, R.; Chen, S.; et al. Osa-miR164a targetsOsNAC60and negatively regulates rice immunity against the blast fungusMagnaporthe oryzae. Plant J. 2018, 95, 584–597. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Wang, K.; Liu, Y.; Chen, Y.; Chen, P.; Shi, Z.; Luo, J.; Jiang, D.; Fan, F.; Zhu, Y.; et al. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2015, 2, 15196. [Google Scholar] [CrossRef]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W.; Deng, Q.; et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef]
- Swetha, C.; Basu, D.; Pachamuthu, K.; Tirumalai, V.; Nair, A.; Prasad, M.; Shivaprasad, P.V. Major Domestication-Related Phenotypes in Indica Rice Are Due to Loss of miRNA-Mediated Laccase Silencing. Plant Cell 2018, 30, 2649–2662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, K.; Xiao, H.; Sun, Y.; Ding, S.; Situ, G.; Li, F. Transgenic microRNA-14 rice shows high resistance to rice stem borer. Plant Biotechnol. J. 2018, 17, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Cuppen, E.; Plasterk, R.H.A. Approaches to microRNA discovery. Nat. Genet. 2006, 38, S2–S7. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, N.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Boil. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef]
- Yang, X.; Li, L. miRDeep-P: A computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 2011, 27, 2614–2615. [Google Scholar] [CrossRef]
- An, J.; Lai, J.; Lehman, M.L.; Nelson, C.C. miRDeep: An integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res. 2012, 41, 727–737. [Google Scholar] [CrossRef]
- Morgado, L.; Johannes, F. Computational tools for plant small RNA detection and categorization. Brief. Bioinform. 2019, 20, 1181–1192. [Google Scholar] [CrossRef] [Green Version]
- Xue, C.; Li, F.; He, T.; Liu, G.-P.; Li, Y.; Zhang, X. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinform. 2005, 6, 310. [Google Scholar] [CrossRef] [Green Version]
- Batuwita, R.; Palade, V. microPred: Effective classification of pre-miRNAs for human miRNA gene prediction. Bioinformatics 2009, 25, 989–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousef, M.; Nebozhyn, M.; Shatkay, H.; Kanterakis, S.; Showe, L.C.; Showe, M.K. Combining multi-species genomic data for microRNA identification using a Naive Bayes classifier. Bioinformatics 2006, 22, 1325–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, D.T.-H.; Wang, C.-C.; Chen, J.-W. Using a kernel density estimation based classifier to predict species-specific microRNA precursors. BMC Bioinform. 2008, 9, S2. [Google Scholar] [CrossRef] [Green Version]
- Xuan, P.; Guo, M.; Liu, X.; Huang, Y.; Li, W. PlantMiRNAPred: Efficient classification of real and pseudo plant pre-miRNAs. Bioinformatics 2011, 27, 1368–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gudyś, A.; Szcześniak, M.; Sikora, M.; Makałowska, I. HuntMi: An efficient and taxon-specific approach in pre-miRNA identification. BMC Bioinform. 2013, 14, 83. [Google Scholar] [CrossRef] [Green Version]
- Williams, P.H.; Eyles, R.; Weiller, G. Plant MicroRNA Prediction by Supervised Machine Learning Using C5.0 Decision Trees. J. Nucleic Acids 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013, 42, D68–D73. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yang, Y.; Zhang, H.; Jiang, X.; Xu, B.; Xue, Y.; Cao, Y.; Zhai, Q.; Zhai, Y.; Xu, M.; et al. Prediction of novel pre-microRNAs with high accuracy through boosting and SVM. Bioinformatics 2011, 27, 1436–1437. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Ma, C.; Deng, H.; Liu, Q.; Zhang, J.; Yi, M. PlantMirP: An efficient computational program for the prediction of plant pre-miRNA by incorporating knowledge-based energy features. Mol. BioSyst. 2016, 12, 3124–3131. [Google Scholar] [CrossRef]
- Zhao, Q.; Yang, Y.; Ren, G.; Ge, E.; Fan, C. Integrating Bipartite Network Projection and KATZ Measure to Identify Novel CircRNA-Disease Associations. IEEE Trans. NanoBiosci. 2019, 18, 578–584. [Google Scholar] [CrossRef]
- Liu, H.; Ren, G.; Chen, H.; Liu, Q.; Yang, Y.; Zhao, Q. Predicting lncRNA–miRNA interactions based on logistic matrix factorization with neighborhood regularized. Knowl. Based Syst. 2020, 191, 105261. [Google Scholar] [CrossRef]
- Liu, Z.; Ren, J.; Cao, J.; He, J.; Yao, X.; Jin, C.; Xue, Y. Systematic analysis of the Plk-mediated phosphoregulation in eukaryotes. Brief. Bioinform. 2012, 14, 344–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Ma, L.; Jia, Q.; Deng, W.; Liu, Z.; Zhang, Y.; Ren, J.; Xue, Y.; Jia, H.; Yang, Q. Systematic characterization of small RNAome during zebrafish early developmental stages. BMC Genom. 2014, 15, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| No. | Feature | Description | Origin |
|---|---|---|---|
| 1–34 | Energy score 1 | Obtained from position-dependent potentials with character pair w. | Novel |
| 35 | Energy score 2 | Obtained from distance-dependent potentials with 3-mer pairs. | plantMirP |
| 36–45 | Ratio of Unpaired bases in sub-region | The secondary structure is divided into 10 parts, and the ratio in each part is calculated. | plantMirP |
| 46 | Size of biggest bulge | A bulge contains at least three adjacent unpaired bases. | plantMirP |
| 47 | n_loops/L | n_loops denotes the number of loops, L is the length of sequence. | plantMirP |
| 48 | n_stems/L | A stem consists of at least three continuous paired bases. | plantMirP |
| 49 | %(|G| + |C|) | (|G| + |C|)/L * 100, here |X| denotes the number of X in sequence. | miPred |
| 50–65 | %XY | |XY|/(L − 1) * 100, |XY| is number of dinucleotide XY in sequence. | miPred |
| 66 | dG | MFE/L, MFE is minimum of free energy of the secondary structure. | miPred |
| 67 | MFE1 | (MFE/L)/%(|G| + |C|) | miPred |
| 68 | MFE2 | (MFE/L)/n_stems | miPred |
| 69 | dP = tot_bases/L | tot_bases is number of base pairs in the secondary structure. | miPred |
| 70 | MFE3 | (MFE/L)/n_loops | microPred |
| 71–73 | |X − Y|/L | |X − Y| is the number of base pairs, (X − Y)∈[(A − U), (G − C), (G − U)] | microPred |
| 74 | Avg_bp_stem | tot_bases/n_stems, n_stems denotes the number of stems. | microPred |
| 75–77 | %(X − Y)/n_stems | %(X − Y) = |X − Y|/tot_bases | microPred |
| 78 | pb/nb | The ratio of paired nucleotides to unpaired nucleotides. | miRD |
| 79 | MCPN | Maximum of consecutive paired nucleotides. | ZmirP [33] |
| 80 | n_bulges/L | n_bulges is the total number of bulges in the secondary structure. | ZmirP |
| 81 | Avg_bp_stem | The ratio of number of base pairs to n_stems. | ZmirP |
| 82 | MFE4 | dG/tot_bases | ZmirP |
| 83 | MFE5 | dG/n_bulges | ZmirP |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Wang, H.; Yao, Y.; Yi, M. PlantMirP-Rice: An Efficient Program for Rice Pre-miRNA Prediction. Genes 2020, 11, 662. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11060662
Zhang H, Wang H, Yao Y, Yi M. PlantMirP-Rice: An Efficient Program for Rice Pre-miRNA Prediction. Genes. 2020; 11(6):662. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11060662
Chicago/Turabian StyleZhang, Huiyu, Hua Wang, Yuangen Yao, and Ming Yi. 2020. "PlantMirP-Rice: An Efficient Program for Rice Pre-miRNA Prediction" Genes 11, no. 6: 662. https://0-doi-org.brum.beds.ac.uk/10.3390/genes11060662




