Gene Flow and Genetic Structure Reveal Reduced Diversity between Generations of a Tropical Tree, Manilkara multifida Penn., in Atlantic Forest Fragments
Abstract
:1. Introduction
2. Materials and Methods
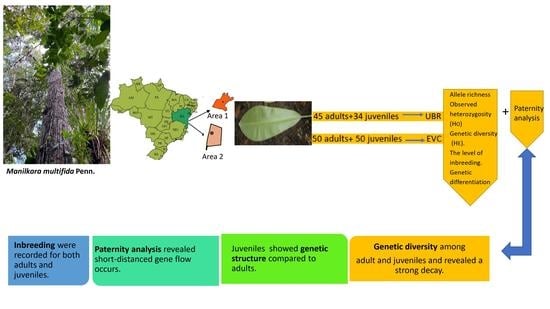
2.1. Study Areas and Tree Sampling
2.2. Genetic Analysis
3. Results
4. Discussion
5. Conclusions and Conservation Implications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dirzo, R.; Raven, P.H. Global State of Biodiversity and Loss. Annu. Rev. Environ. Resour. 2003, 28, 137–167. [Google Scholar] [CrossRef] [Green Version]
- Phillips, H.R.P.; Newbold, T.; Purvis, A. Land-use effects on local biodiversity in tropical forests vary between continents. Biodivers. Conserv. 2017, 26, 2251–2270. [Google Scholar] [CrossRef] [Green Version]
- Laurance, W.F.; Sayer, J.; Cassman, K.G. Agricultural expansion and its impacts on tropical nature. Trends Ecol. Evol. 2014, 29, 107–116. [Google Scholar] [CrossRef]
- Taubert, F.; Fischer, R.; Groeneveld, J.; Lehmann, S.; Müller, M.S.; Rödig, E.; Wiegand, T.; Huth, A. Global patterns of tropical forest fragmentation. Nature 2018, 554, 519–522. [Google Scholar] [CrossRef]
- Betts, M.G.; Wolf, C.; Pfeifer, M.; Banks-Leite, C.; Arroyo-Rodríguez, V.; Ribeiro, D.B.; Barlow, J.; Eigenbrod, F.; Faria, D.; Fletcher, R.J.; et al. Extinction filters mediate the global effects of habitat fragmentation on animals. Science 2019, 366, 1236–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sork, V.L.; Smouse, P.E. Genetic analysis of landscape connectivity in tree populations. Landsc. Ecol. 2006, 21, 821–836. [Google Scholar] [CrossRef]
- Kremer, A.; Ronce, O.; Robledo-Arnuncio, J.J.; Guillaume, F.; Bohrer, G.; Nathan, R.; Bridle, J.R.; Gomulkiewicz, R.; Klein, E.K.; Ritland, K.; et al. Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecol. Lett. 2012, 15, 378–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mijangos, J.L.; Pacioni, C.; Spencer, P.B.S.; Craig, M.D. Contribution of genetics to ecological restoration. Mol. Ecol. 2015, 24, 22–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, E.R.; Barker, C.; Powell, I.; Ashton, P.A. Genetic diversity and connectivity in fragmented populations of Rhinanthus minor in two regions with contrasting land-use. Biodivers. Conserv. 2019, 28, 3159–3181. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, C.D.S.; Ballesteros-Mejia, L.; Ribeiro, M.C.; Côrtes, M.C.; Santos, A.S.; Collevatti, R.G. Climatic stability and contemporary human impacts affect the genetic diversity and conservation status of a tropical palm in the Atlantic Forest of Brazil. Conserv. Genet. 2017, 18, 467–478. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira Buzatti, R.S.; Ribeiro, R.A.; de Lemos Filho, J.P.; Lovato, M.B. Fine-scale spatial genetic structure of Dalbergia nigra (Fabaceae), a threatened and endemic tree of the Brazilian Atlantic Forest. Genet. Mol. Biol. 2012, 35, 838–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahrig, L. Effects of Habitat Fragmentation on Biodiversity. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 487–515. [Google Scholar] [CrossRef] [Green Version]
- Ouborg, N.J.; Pertoldi, C.; Loeschcke, V.; Bijlsma, R.; Hedrick, P.W. Conservation genetics in transition to conservation genomics. Trends Genet. 2010, 26, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Holderegger, R.; Kamm, U.; Gugerli, F. Adaptive vs. neutral genetic diversity: Implications for landscape genetics. Landsc. Ecol. 2006, 21, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Keller, L.F.; Biebach, I.; Ewing, S.R.; Hoeck, P.E.A. The genetics of reintroductions: Inbreeding and genetic drift. In Reintroduction Biology; John Wiley & Sons: Hoboken, NJ, USA, 2012; pp. 360–394. [Google Scholar]
- Vellend, M.; Geber, M.A. Connections between species diversity and genetic diversity. Ecol. Lett. 2005, 8, 767–781. [Google Scholar] [CrossRef]
- Gaston, K.J. Global patterns in biodiversity. Nature 2000, 405, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Willi, Y. Detecting genetic responses to environmental change. Nat. Rev. Genet. 2008, 9, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Endler, J.A. Natural Selection in the Wild. (MPB-21); Princeton University Press: Princeton, NJ, USA, 1986; Volume 21. [Google Scholar]
- Buffalo, V.; Coop, G. Estimating the genome-wide contribution of selection to temporal allele frequency change. Proc. Natl. Acad. Sci. USA 2020, 117, 20672. [Google Scholar] [CrossRef]
- Chen, N.; Juric, I.; Cosgrove, E.J.; Bowman, R.; Fitzpatrick, J.W.; Schoech, S.J.; Clark, A.G.; Coop, G. Allele frequency dynamics in a pedigreed natural population. Proc. Natl. Acad. Sci. USA 2019, 116, 2158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, D.; Slate, J. Understanding genetic changes between generations. Proc. Natl. Acad. Sci. USA 2019, 116, 1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martini, A.M.Z.; Fiaschi, P.; Amorim, A.M.; Paixão, J.L.D. A hot-point within a hot-spot: A high diversity site in Brazil’s Atlantic Forest. Biodivers. Conserv. 2007, 16, 3111–3128. [Google Scholar] [CrossRef]
- Landau, E.; Hirsch, A.; Musinsky, J. Vegetation cover and land use in the Atlantic Coastal Forest of Southern Bahia, Brazil, based on satellite imagery: A comparison among municipalities. Mem. N. Y. Bot. Gard. 2008, 100, 221–244. [Google Scholar]
- Schroth, G.; Faria, D.; Araujo, M.; Bede, L.; van Bael, S.A.; Cassano, C.R.; Oliveira, L.C.; Delabie, J.H.C. Conservation in tropical landscape mosaics: The case of the cacao landscape of southern Bahia, Brazil. Biodivers. Conserv. 2011, 20, 1635–1654. [Google Scholar] [CrossRef]
- Borges, D.B.; Mariano-Neto, E.; Gaiotto, F.A. Development of microsatellite primers for Melanoxylon brauna (Fabaceae): An endangered and endemic tree from the Brazilian Atlantic Forest. Conserv. Genet. Resour. 2015, 7, 65–68. [Google Scholar] [CrossRef]
- Borges, L.A.; Sobrinho, M.S.; Lopes, A.V. Phenology, pollination, and breeding system of the threatened tree Caesalpinia echinata Lam. (Fabaceae), and a review of studies on the reproductive biology in the genus. Flora Morphol. Distrib. Funct. Ecol. Plants 2009, 204, 111–130. [Google Scholar] [CrossRef]
- Gibson, J.P.; Rice, S.A.; Stucke, C.M. Comparison of population genetic diversity between a rare, narrowly distributed species and a common, widespread species of Alnus (Betulaceae). Am. J. Bot. 2008, 95, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Coelho, N.; Gonçalves, S.; Romano, A. Endemic Plant Species Conservation: Biotechnological Approaches. Plants 2020, 9, 345. [Google Scholar] [CrossRef] [Green Version]
- Turchetto, C.; Segatto, A.L.A.; Mäder, G.; Rodrigues, D.M.; Bonatto, S.L.; Freitas, L.B. High levels of genetic diversity and population structure in an endemic and rare species: Implications for conservation. AoB Plants 2016, 8, plw002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isik, K. Rare and endemic species: Why are they prone to extinction? Turk. J. Bot. 2011, 35, 411–417. [Google Scholar]
- Foggi, B.; Viciani, D.; Baldini, R.M.; Carta, A.; Guidi, T. Conservation assessment of the endemic plants of the Tuscan Archipelago, Italy. Oryx 2014, 49, 118–126. [Google Scholar] [CrossRef] [Green Version]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. N. For. 1992, 6, 95–124. [Google Scholar]
- Furches, M.S.; Small, R.L.; Furches, A. Genetic diversity in three endangered pitcher plant species (Sarracenia; Sarraceniaceae) is lower than widespread congeners. Am. J. Bot. 2013, 100, 2092–2101. [Google Scholar] [CrossRef] [PubMed]
- Bax, V.; Francesconi, W. Conservation gaps and priorities in the Tropical Andes biodiversity hotspot: Implications for the expansion of protected areas. J. Environ. Manag. 2019, 232, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Waldron, A.; Mooers, A.O.; Miller, D.C.; Nibbelink, N.; Redding, D.; Kuhn, T.S.; Roberts, J.T.; Gittleman, J.L. Targeting global conservation funding to limit immediate biodiversity declines. Proc. Natl. Acad. Sci. USA 2013, 110, 12144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, L.C.; Hankerson, S.J.; Dietz, J.M.; Raboy, B.E. Key tree species for the golden-headed lion tamarin and implications for shade-cocoa management in southern Bahia, Brazil. Anim. Conserv. 2010, 13, 60–70. [Google Scholar] [CrossRef]
- Amorim, E.; Martinelli, G.; Gomes, M. Manilkara multifida; The IUCN Red List of Threatened Species 2020: e.T35616A176126685; IUCN: Gland, Switzerland, 2020. [Google Scholar] [CrossRef]
- Ribeiro, M.C.; Metzger, J.P.; Martensen, A.C.; Ponzoni, F.J.; Hirota, M.M. The Brazilian Atlantic Forest: How much is left, and how is the remaining forest distributed? Implications for conservation. Biol. Conserv. 2009, 142, 1141–1153. [Google Scholar] [CrossRef]
- Thomas, W. Natural Vegetation Types in Southern Bahia; Instituto de Estudos Sócio-Ambientais do Sul da Bahia e Conservation International do Brasil: Ilheus, Brazil, 2003; Volume 2. [Google Scholar]
- Schiavetti, A.; Fonseca, M.; Bedê, L.; Pinto, L.P. Plano de Manejo: Reserva Particular do Patrimônio Natural Estação Veracel; Conservação Internacional & Instituto Bioatlântica: Rio de Janeiro, Brazil, 2007; Volume 14, p. 293. [Google Scholar]
- Cassano, C.R.; Schroth, G.; Faria, D.; Delabie, J.H.C.; Bede, L. Landscape and farm scale management to enhance biodiversity conservation in the cocoa producing region of southern Bahia, Brazil. Biodivers. Conserv. 2009, 18, 577–603. [Google Scholar] [CrossRef]
- Azevedo, V.C.R.; Kanashiro, M.; Ciampi, A.Y.; Grattapaglia, D. Genetic Structure and Mating System of Manilkara huberi (Ducke) A. Chev., a Heavily Logged Amazonian Timber Species. J. Hered. 2007, 98, 646–654. [Google Scholar] [CrossRef] [Green Version]
- Doyle, J. DNA protocols for plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. [Google Scholar]
- Moraes, R.C.S.; Vivas, C.V.; Oliveira, F.A.; Menezes, I.P.P.; van den Berg, C.; Gaiotto, F.A. Microsatellite markers for an endemic Atlantic Forest tree, Manilkara multifida (Sapotaceae). AoB Plants 2013, 5, plt006. [Google Scholar] [CrossRef] [Green Version]
- Goudet, J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Marshall, T.C.; Slate, J.; Kruuk, L.E.B.; Pemberton, J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardy, O.J.; Vekemans, X. Spagedi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef] [Green Version]
- Loiselle, B.A.; Sork, V.L.; Nason, J.; Graham, C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (RuBIACEAE). Am. J. Bot. 1995, 82, 1420–1425. [Google Scholar] [CrossRef]
- Greenbaum, G.; Templeton, A.R.; Zarmi, Y.; Bar-David, S. Allelic Richness following Population Founding Events—A Stochastic Modeling Framework Incorporating Gene Flow and Genetic Drift. PLoS ONE 2014, 9, e115203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dantas, L.G.; Esposito, T.; de Sousa, A.C.B.; Félix, L.; Amorim, L.L.B.; Benko-Iseppon, A.M.; Batalha-Filho, H.; Pedrosa-Harand, A. Low genetic diversity and high differentiation among relict populations of the neotropical gymnosperm Podocarpus sellowii (Klotz.) in the Atlantic Forest. Genetica 2015, 143, 21–30. [Google Scholar] [CrossRef]
- Fundação SOS Mata Atlântica; INPE. Atlas dos Remanescentes Florestais da Mata Atlântica: Período 2019/2020, Relatório Técnico; Fundação SOS Mata Atlântica: São Paulo, Brazil, 2021; p. 73.
- Collevatti, R.G.; Grattapaglia, D.; Hay, J.D. Population genetic structure of the endangered tropical tree species Caryocar brasiliense, based on variability at microsatellite loci. Mol. Ecol. 2001, 10, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.T.; Ison, J.L.; Ashley, M.V.; Howe, H.F. The Paradox of Forest Fragmentation Genetics. Conserv. Biol. 2008, 22, 878–885. [Google Scholar] [CrossRef]
- Da Silva Carvalho, C.; Ribeiro, M.C.; Côrtes, M.C.; Galetti, M.; Collevatti, R.G. Contemporary and historic factors influence differently genetic differentiation and diversity in a tropical palm. Heredity 2015, 115, 216–224. [Google Scholar] [CrossRef] [Green Version]
- Almeida-Rocha, J.M.; Soares, L.A.S.S.; Andrade, E.R.; Gaiotto, F.A.; Cazetta, E. The impact of anthropogenic disturbances on the genetic diversity of terrestrial species: A global meta-analysis. Mol. Ecol. 2020, 29, 4812–4822. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. The theoretical population genetics of variable selection and migration. Annu. Rev. Genet. 1976, 10, 253–280. [Google Scholar] [CrossRef]
- Browne, L.; Ottewell, K.; Karubian, J. Short-term genetic consequences of habitat loss and fragmentation for the neotropical palm Oenocarpus bataua. Heredity 2015, 115, 389–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farwig, N.; Braun, C.; Böhning-Gaese, K. Human disturbance reduces genetic diversity of an endangered tropical tree, Prunus africana (Rosaceae). Conserv. Genet. 2007, 9, 317. [Google Scholar] [CrossRef]
- André, T.; Lemes, M.R.; Grogan, J.; Gribel, R. Post-logging loss of genetic diversity in a mahogany (Swietenia macrophylla King, Meliaceae) population in Brazilian Amazonia. For. Ecol. Manag. 2008, 255, 340–345. [Google Scholar] [CrossRef]
- Maués, M.M. Sistemas de polinização no dossel de uma florestaombrófila densa na Amazônia. In Proceedings of the VIII Congresso de Ecologia do Brasil; Sociedade de Ecologia do Brasil: Caxambu, Brazil, 2007. [Google Scholar]
- Gonçalves, R.; Brandão, C. Diversity of bees (Hymenoptera, Apidae) along a latitudinal gradient in the Atlantic Forest. Biota Neotropica 2008, 8. [Google Scholar] [CrossRef]
- Angers, B.; Estoup, A.; Jarne, P. Microsatellite Size Homoplasy, SSCP, and Population Structure: A Case Study in the Freshwater Snail Bulinus truncatus. Mol. Biol. Evol. 2000, 17, 1926–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnaval, A.C.; Moritz, C. Historical climate modelling predicts patterns of current biodiversity in the Brazilian Atlantic forest. J. Biogeogr. 2008, 35, 1187–1201. [Google Scholar] [CrossRef]
- Konzen, E.; Martins, M. Contrasting levels of genetic diversity among populations of the endangered tropical palm Euterpe edulis martius. Cerne 2017, 23, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Reed, D.H.; Frankham, R. Correlation between Fitness and Genetic Diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Schlaepfer, D.R.; Braschler, B.; Rusterholz, H.-P.; Baur, B. Genetic effects of anthropogenic habitat fragmentation on remnant animal and plant populations: A meta-analysis. Ecosphere 2018, 9, e02488. [Google Scholar] [CrossRef]
- Sebbenn, A.M.; Carvalho, A.C.M.; Freitas, M.L.M.; Moraes, S.M.B.; Gaino, A.P.S.C.; da Silva, J.M.; Jolivet, C.; Moraes, M.L.T. Low levels of realized seed and pollen gene flow and strong spatial genetic structure in a small, isolated and fragmented population of the tropical tree Copaifera langsdorffii Desf. Heredity 2011, 106, 134–145. [Google Scholar] [CrossRef] [Green Version]
- Garcia, C.; Jordano, P.; Godoy, J. Contemporary pollen and seed dispersal in a Prunus mahaleb population: Patterns in distance and direction. Mol. Ecol. 2007, 16, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Bittencourt, J.V.M.; Sebbenn, A.M. Patterns of pollen and seed dispersal in a small, fragmented population of the wind-pollinated tree Araucaria angustifolia in southern Brazil. Heredity 2007, 99, 580–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, L.A.S.S.; Cazetta, E.; Santos, L.R.; França, D.d.S.; Gaiotto, F.A. Anthropogenic Disturbances Eroding the Genetic Diversity of a Threatened Palm Tree: A Multiscale Approach. Front. Genet. 2019, 10, 1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Site | Samples | Na | Ar | HE | HO | f | Θ | |
|---|---|---|---|---|---|---|---|---|
| Adults | EVC | 50 | 12.9 | 9.6 | 0.815 | 0.720 | 0.103 ** | 0.017 * |
| UBR | 45 | 10.9 | 8.8 | 0.818 | 0.736 | 0.097 ** | ||
| Juveniles | EVC | 50 | 9.9 | 8.1 | 0.731 | 0.463 | 0.366 ** | 0.109 *** |
| UBR | 33 | 8.1 | 7.1 | 0.729 | 0.560 | 0.231 ** |
| Juvenile ID | Parent 1 ID | Parent 2 ID | LOD Score | TRIO Confidence | Distance Juveniles-P1 (km) | Distance Juveniles-P2 (km) | Distance P1-P2 (km) |
|---|---|---|---|---|---|---|---|
| EVC-J08 | EVC-A43 | EVC-A43 | 3.96 | * | 4.81 | - | - |
| UBR-J11 | UBR-A35 | UBR-A35 | 7.54 | * | 13.0 | - | - |
| EVC-J15 | UBR-A32 | UBR-A32 | 4.50 | * | 127.64 | - | - |
| UBR-J30 | UBR-A43 | UBR-A43 | 2.90 | * | - | - | - |
| UBR-J34 | EVC-A113 | UBR-A31 | 3.81 | * | - | - | - |
| EVC-J35 | EVC-A21 | EVC-A21 | 4.51 | * | 5.28 | - | - |
| EVC-J45 | EVC-A39 | EVC-A21 | 2.99 | * | 0.21 | 5.24 | 5.03 |
| EVC-J02 | EVC-A122 | EVC-A30 | 1.43 | + | - | 5.19 | - |
| EVC-J04 | EVC-A43 | EVC-A43 | 2.15 | + | 4.92 | - | - |
| EVC-J13 | EVC-A118 | UBR-A40 | 2.35 × 10−1 | + | - | - | - |
| UBR-J15 | UBR-A45 | UBR-A46 | 2.26 | + | - | - | - |
| EVC-J16 | EVC-A118 | UBR-A23 | 8.33 × 10−1 | + | - | 127.58 | - |
| UBR-J16 | EVC-A28 | EVC-A28 | 5.98 × 10−1 | + | 123.48 | - | - |
| EVC-J22 | UBR-A33 | EVC-A118 | 2.19 | + | 127.46 | - | - |
| UBR-J22 | UBR-A46 | EVC-A122 | 8.39 × 10−2 | + | - | - | - |
| EVC-J29 | UBR-A37 | EVC-A22 | 2.31 × 10−1 | + | - | 5.28 | - |
| EVC-J30 | EVC-A59 | UBR-A43 | 3.82 × 10−1 | + | - | - | - |
| UBR-J31 | EVC-A122 | EVC-A113 | 7.59 × 10−1 | + | - | - | - |
| EVC-J32 | UBR-A28 | UBR-A34 | 1.76 | + | 124 | - | - |
| EVC-J43 | EVC-A21 | EVC-A21 | 1.78 | + | 5.24 | - | - |
| EVC-J49 | EVC-A122 | UBR-A26 | 1.70 | + | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waqar, Z.; Moraes, R.C.S.; Benchimol, M.; Morante-Filho, J.C.; Mariano-Neto, E.; Gaiotto, F.A. Gene Flow and Genetic Structure Reveal Reduced Diversity between Generations of a Tropical Tree, Manilkara multifida Penn., in Atlantic Forest Fragments. Genes 2021, 12, 2025. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12122025
Waqar Z, Moraes RCS, Benchimol M, Morante-Filho JC, Mariano-Neto E, Gaiotto FA. Gene Flow and Genetic Structure Reveal Reduced Diversity between Generations of a Tropical Tree, Manilkara multifida Penn., in Atlantic Forest Fragments. Genes. 2021; 12(12):2025. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12122025
Chicago/Turabian StyleWaqar, Zubaria, Ramiris César Souza Moraes, Maíra Benchimol, José Carlos Morante-Filho, Eduardo Mariano-Neto, and Fernanda Amato Gaiotto. 2021. "Gene Flow and Genetic Structure Reveal Reduced Diversity between Generations of a Tropical Tree, Manilkara multifida Penn., in Atlantic Forest Fragments" Genes 12, no. 12: 2025. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12122025