Genomic Regions Associated with Variation in Pigmentation Loss in Saddle Tan Beagles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Phenotyping and Genotyping
2.3. GWAS
3. Results
3.1. Color Phenotype
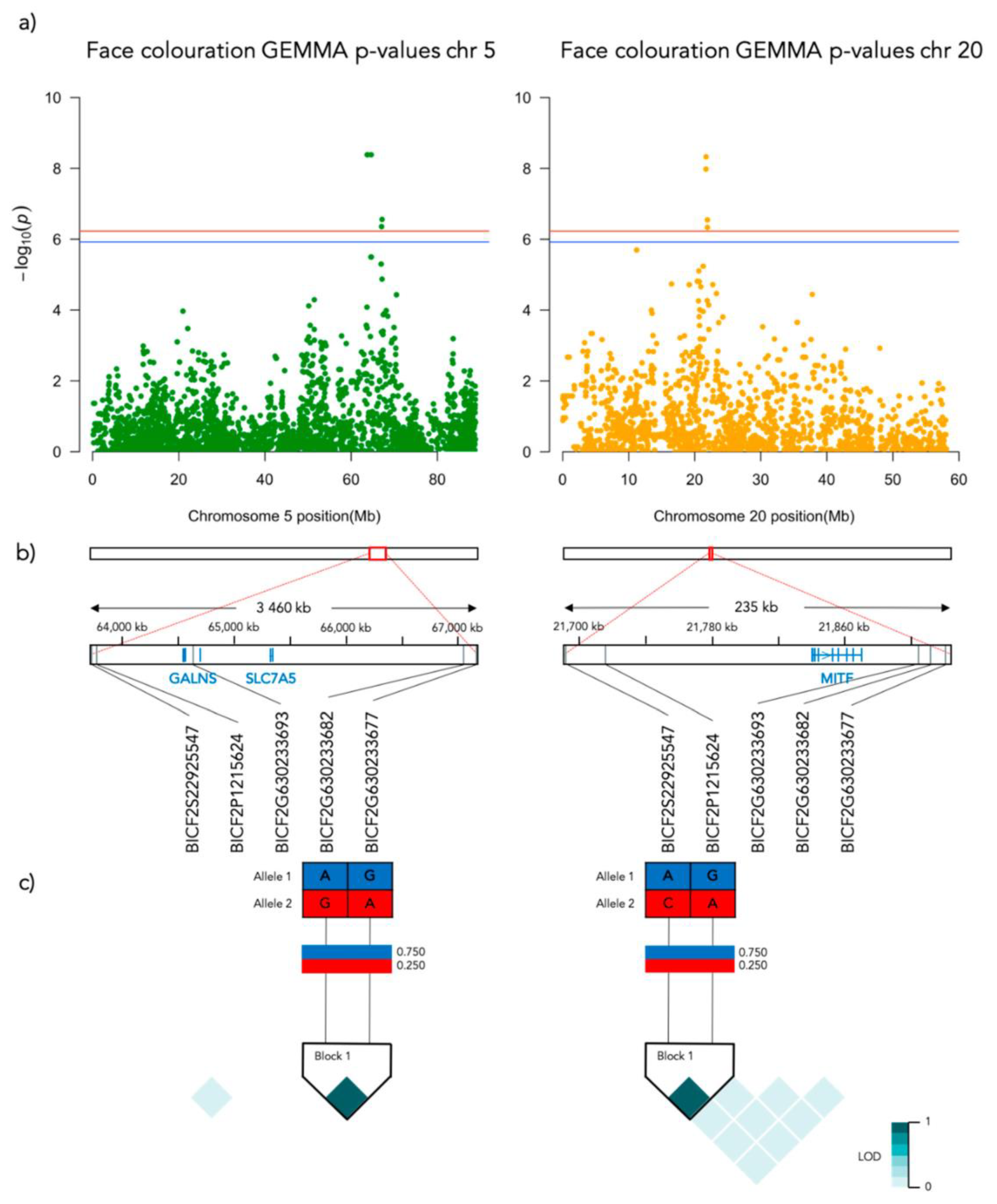
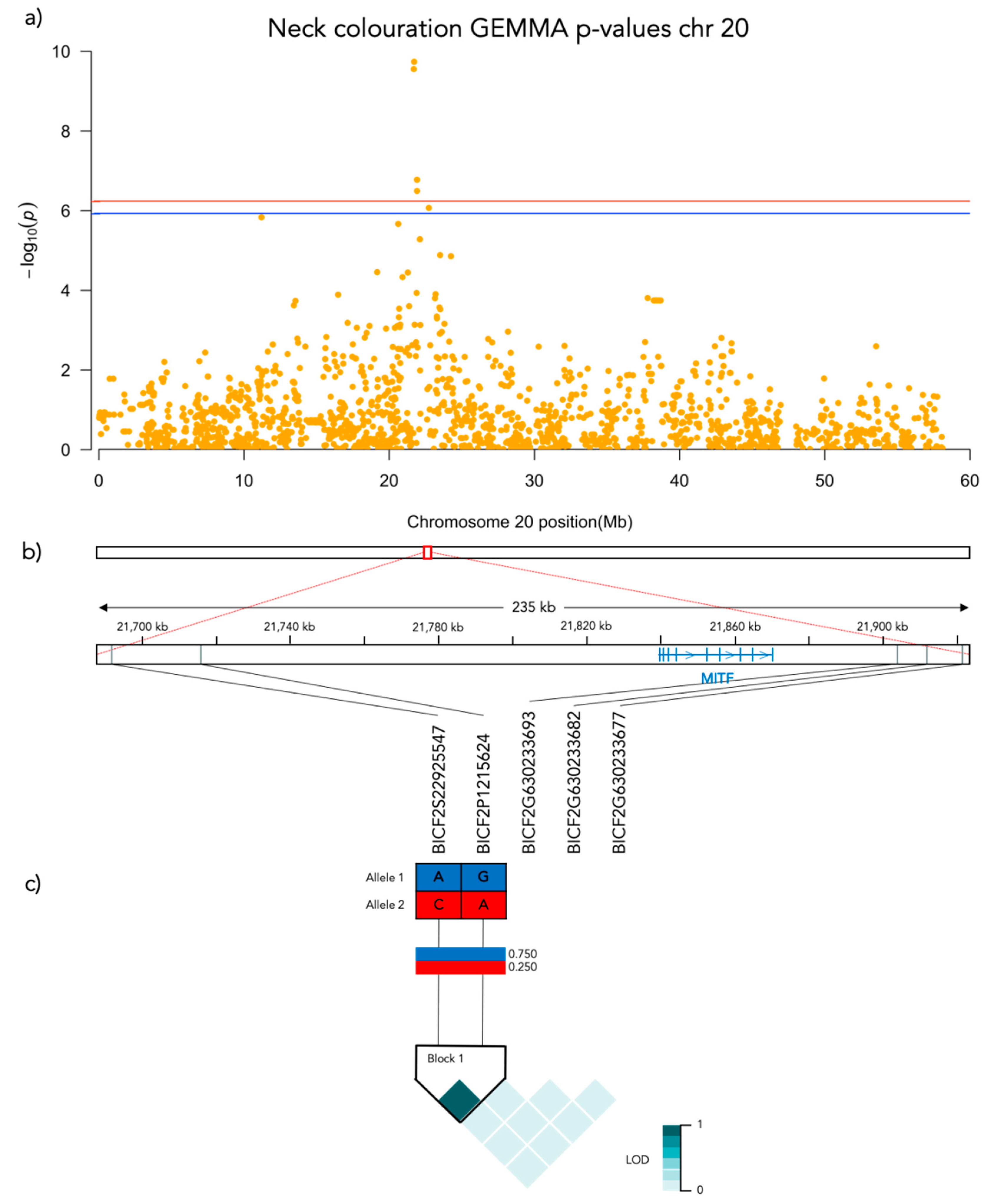
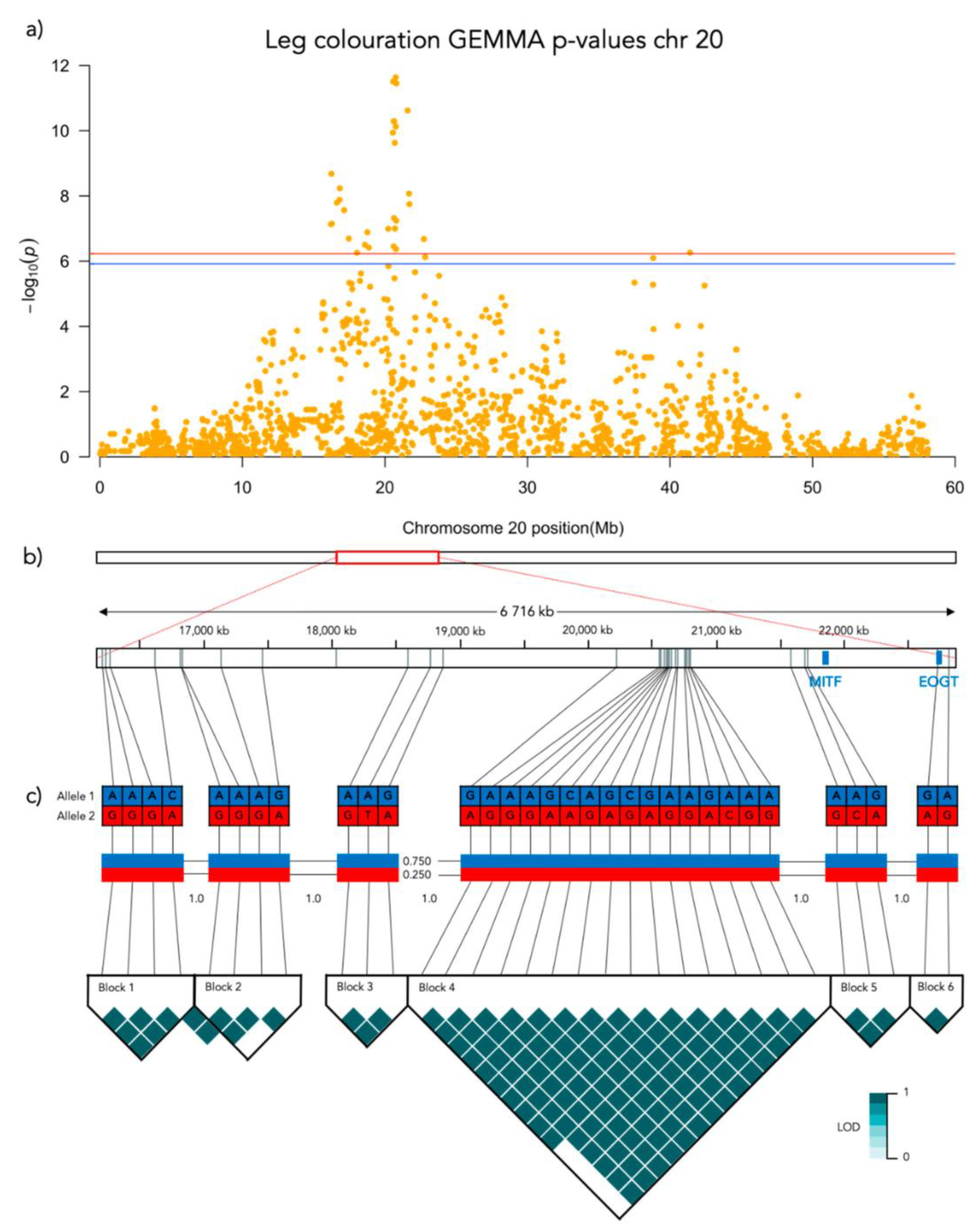
3.2. GWAS
3.3. Genotype Associations
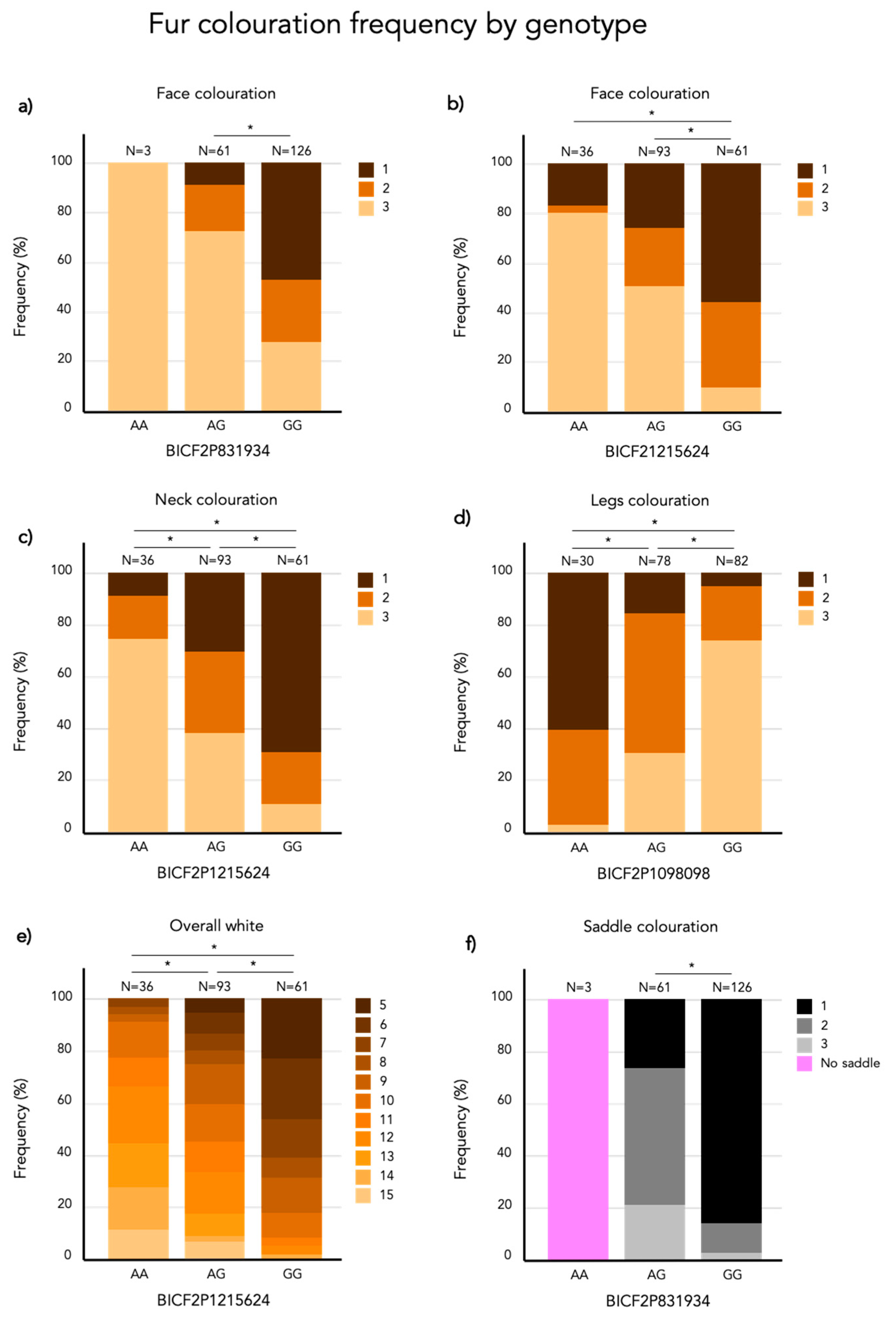
3.3.1. Face Coloration
3.3.2. Neck Coloration
3.3.3. Leg Coloration
3.3.4. Overall White
3.3.5. Saddle Coloration
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Darwin, C. The Variation of Animals and Plants under Domestication, Authorized ed.; O. Judd & Company: New York, NY, USA, 1868. [Google Scholar]
- Trut, L.N. Early canid domestication: The farm-fox experiment. Am. Sci. 1999, 87, 160–169. [Google Scholar] [CrossRef]
- Karlsson, A.-C.; Kerje, S.; Andersson, L.; Jensén, P. Genotype at thePMEL17locus affects social and explorative behaviour in chickens. Br. Poult. Sci. 2010, 51, 170–177. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J., III; Zody, M.C.; et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef] [PubMed]
- Ollivier, M.; Tresset, A.; Hitte, C.; Petit, C.; Hughes, S.; Gillet, B.; Duffraisse, M.; Pionnier-Capitan, M.; Lagoutte, L.; Arbogast, R.-M.; et al. Evidence of coat color variation sheds new llight on ancient canids. PLoS ONE 2013, 8, e75110. [Google Scholar] [CrossRef]
- Kaelin, C.B.; Barsh, G.S. Molecular Genetics of Coat Color, Texture and Length in the Dog in the Genetics of the Dog; CABI: Wallingford, UK; Cambridge, MA, USA, 2012; pp. 57–82. [Google Scholar]
- Schmutz, S.M.; Berryere, T.G. Genes affecting coat color and pattern in domestic dogs: A review. Anim. Genet. 2007, 38, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Saif, R.; Iftekhar, A.; Asif, F.; Alghanem, M.S. Dog coat color genetics: A review. Adv. Life Sci. 2020, 7, 215–224. [Google Scholar]
- Little, C.C. The Inheritance of Coat Color in Dogs; Howell Books: New York, NY, USA, 1957. [Google Scholar]
- Bannasch, D.L.; Kaelin, C.B.; Letko, P.; Loechel, P.; Hug, P.; Jagannathan, P.; Henkel, P.; Roosje, P.; Hytönen, P.K.; Lohi, P.; et al. Dog color patterns explained by modular promoters of ancient canid origin. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dreger, D.L.; Parker, H.G.; Ostrander, E.A.; Schmutz, S.M. Identification of a Mutation that Is Associated with the Saddle Tan and Black-and-Tan Phenotypes in Basset Hounds and Pembroke Welsh Corgis. J. Hered. 2013, 104, 399–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreger, D.L.; Hooser, B.N.; Hughes, A.M.; Ganesan, B.; Donner, J.; Anderson, H.; Holtvoigt, L.; Ekenstedt, K.J. True Colors: Commercially-acquired morphological genotypes reveal hidden allele variation among dog breeds, informing both trait ancestry and breed potential. PLoS ONE 2019, 14, e0223995. [Google Scholar] [CrossRef] [Green Version]
- Körberg, I.B.; Sundström, E.; Meadows, J.R.S.; Pielberg, G.R.; Gustafson, U.; Hedhammar, Å; Karlsson, E.K.; Seddon, J.; Söderberg, A.; Vilà, C.; et al. A Simple Repeat Polymorphism in the MITF-M Promoter Is a Key Regulator of White Spotting in Dogs. PLoS ONE 2014, 9, e104363. [Google Scholar] [CrossRef] [Green Version]
- Dreger, D.L.; Schmutz, S.M. A SINE insertion causes the black-and-tan and saddle tan phenotypes in domestic dogs. J. Hered. 2011, 102 (Suppl. 1), S11–S18. [Google Scholar] [CrossRef]
- Persson, M.E.; Wright, D.; Roth, L.S.V.; Batakis, P.; Jensen, P. Genomic Regions Associated With Interspecies Communication in Dogs Contain Genes Related to Human Social Disorders. Sci. Rep. 2016, 6, 33439. [Google Scholar] [CrossRef] [Green Version]
- Persson, M.E.; Roth, L.S.V.; Johnsson, M.; Wright, D.; Jensen, P. Human-directed social behaviour in dogs shows significant heritability. Genes Brain Behav. 2015, 14, 337–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, S.; Armstrong, L.L.; Bradford, Y.; Carlson, C.S.; Crawford, D.C.; Crenshaw, A.T.; De Andrade, M.; Doheny, K.F.; Haines, J.L.; Hayes, G.; et al. Quality Control Procedures for Genome-Wide Association Studies. Curr. Protoc. Hum. Genet. 2011, 68, 1.19.1–1.19.18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; Van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [Green Version]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothschild, M.F.; Van Cleave, P.S.; Glenn, K.L.; Carlstrom, L.P.; Ellinwood, N.M. Association of MITF with white spotting in Beagle crosses and Newfoundland dogs. Anim. Genet. 2006, 37, 606–607. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, S.M.; Berryere, T.G.; Dreger, D.L. MITF and White Spotting in Dogs: A Population Study. J. Hered. 2009, 100, S66–S74. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, E.K.; Baranowska, I.; Wade, C.M.; Hillbertz, N.H.C.S.; Zody, M.C.; Anderson, N.; Biagi, T.M.; Patterson, N.; Pielberg, G.R.; Kulbokas, E.J.; et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 2007, 39, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Everts, R.E.; Rothuizen, J.; van Oost, B.A. Identification of a premature stop codon in the melanocyte-stimulating hormone receptor gene (MC1R) in Labrador and Golden retrievers with yellow coat color. Anim. Genet. 2000, 31, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Newton, J.; Wilkie, A.L.; He, L.; Jordan, S.A.; Metallinos, D.L.; Holmes, N.G.; Jackson, I.J.; Barsh, G.S. Melanocortin 1 receptor variation in the domestic dog. Mamm. Genome 2000, 11, 24–30. [Google Scholar] [CrossRef]
- Woolf, C.M. Common white facial markings in bay and chestnut Arabian horsses and their hybrids. J. Hered. 1991, 82, 167–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strain, G.M. The Genetics of Deafness in Domestic Animals. Front. Vet. Sci. 2015, 2, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
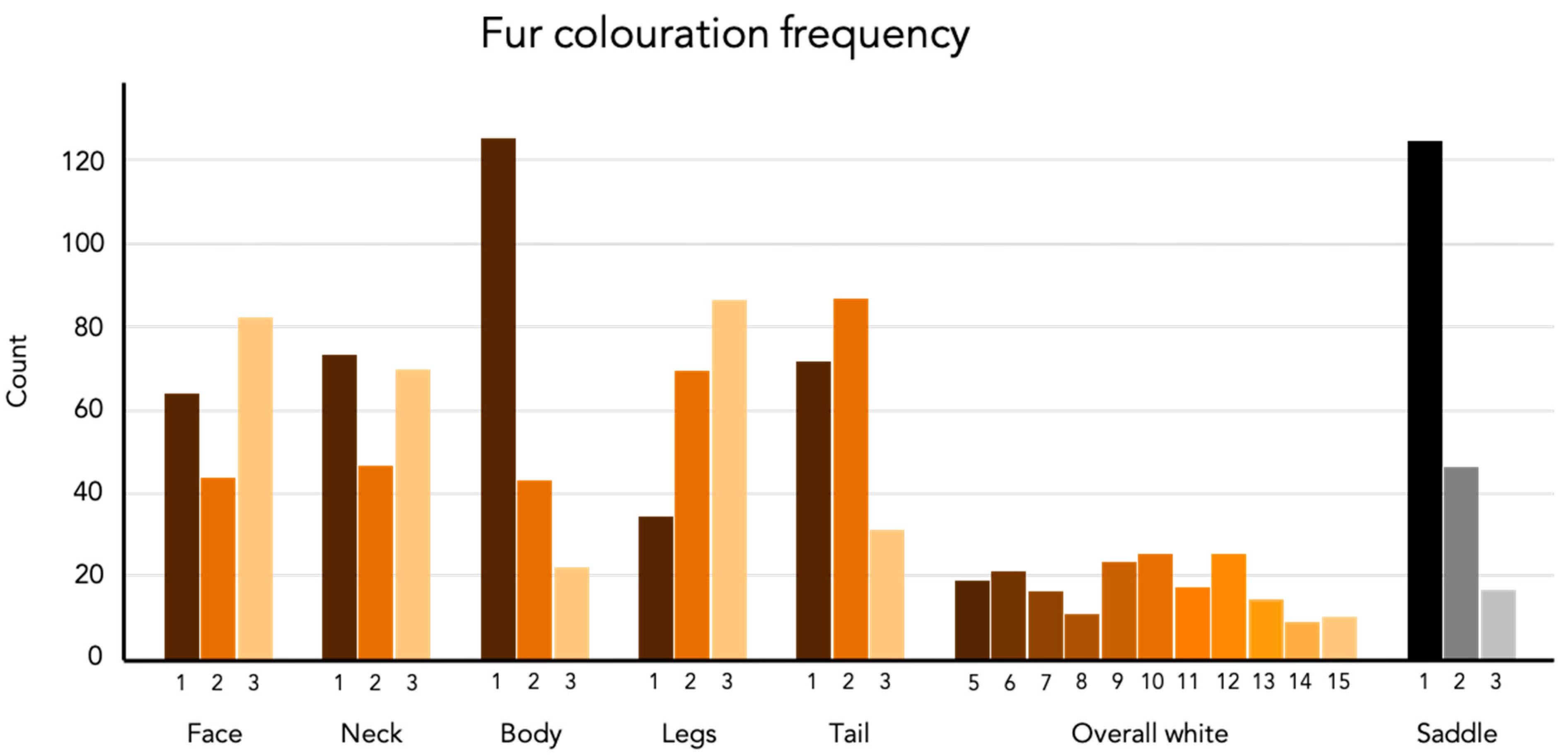




| Phenotype | Score | Name | Description |
|---|---|---|---|
| Face | 1 | No or residual white | Solid colored face with no, or only, residual white around the nose. |
| 2 | White muzzle | White markings around the nose, partly or completely extending over the muzzle, no blaze but a thin streak may occur. | |
| 3 | Blaze | As described above but also including a blaze reaching to or above eye level. | |
| Neck | 1 | No white | Except for the ventral side, solid colored neck. |
| 2 | Residual white | Except for the ventral side, small residual white patches or thin stripes around the neck. | |
| 3 | Collar | The white on the ventral side of the neck extends as a collar towards or around the dorsal side. Large patches. | |
| Body | 1 | Solid | No or only residual white patches on the body except for the neck area and the ventral side. |
| 2 | Some white | Only one or two smaller white patch/patches on the body except for the neck area and the ventral side, usually extending from the hind legs. | |
| 3 | White patches | Large white patches, usually on both sides of the body. | |
| Legs | 1 | Half white | Legs are less white than and does not fulfil the criteria of score 2. |
| 2 | Mostly white | At least 3 legs are between 2 and 3 to fully white. | |
| 3 | Completely white | At least 3 legs fully white and the white extends from the leg on to the body from at least one leg. | |
| Tail | 1 | White tip | 1/4 or less of the tail is white. |
| 2 | Half white | More than 1/4 but less than 3/4 of the tail is white. | |
| 3 | Mostly white | The tail is more than 3/4 white. | |
| Overall white | 5–15 | The sum of scores from face, neck, body, legs and tail. | |
| Saddle | 1 | Solid | Solid black saddle from the neck to the tail base. |
| 2 | Semi-faded | Mostly solid black saddle from the neck to the tail base, but brown patches occurs. | |
| 3 | Faded | No or mostly no solid black saddle. |
| Face | Neck | Body | Legs | Tail | Overall | Saddle | |
|---|---|---|---|---|---|---|---|
| PVE | 0.40 | 0.59 | 0.55 | 0.79 | 0.46 | 0.77 | 0.61 |
| SE (PVE) | 0.13 | 0.15 | 0.14 | 0.11 | 0.14 | 0.11 | 0.19 |
| λ | 1.07 | 1.05 | 1.11 | 1.08 | 1.04 | 1.09 | 1.09 |
| SE (λ) | 0.0004 | 0.0003 | 0.0002 | 0.0009 | 0.0002 | 0.0007 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nord, M.E.; Jensen, P. Genomic Regions Associated with Variation in Pigmentation Loss in Saddle Tan Beagles. Genes 2021, 12, 316. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12020316
Nord ME, Jensen P. Genomic Regions Associated with Variation in Pigmentation Loss in Saddle Tan Beagles. Genes. 2021; 12(2):316. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12020316
Chicago/Turabian StyleNord, Mia E., and Per Jensen. 2021. "Genomic Regions Associated with Variation in Pigmentation Loss in Saddle Tan Beagles" Genes 12, no. 2: 316. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12020316






