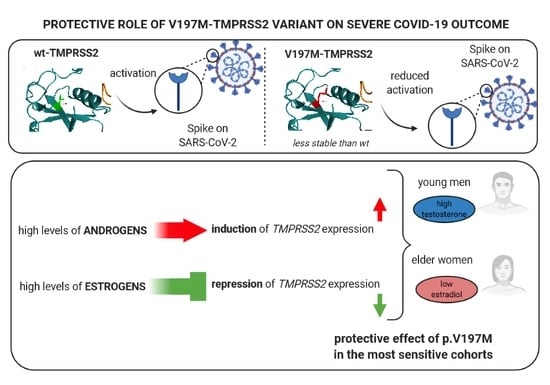
Protective Role of a TMPRSS2 Variant on Severe COVID-19 Outcome in Young Males and Elderly Women
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Lucas, J.M.; Heinlein, C.; Kim, T.; Hernandez, S.A.; Malik, M.S.; True, L.D.; Morrissey, C.; Corey, E.; Montgomery, B.; Mostaghel, E.; et al. The Androgen-Regulated Protease TMPRSS2 Activates a Proteolytic Cascade Involving Components of the Tumor Microenvironment and Promotes Prostate Cancer Metastasis. Cancer Discov. 2014, 4, 1310–1325. [Google Scholar] [CrossRef] [Green Version]
- Wilson, S.; Greer, B.; Hooper, J.; Zijlstra, A.; Walker, B.; Quigley, J.; Hawthorne, S. The Membrane-Anchored Serine Protease, TMPRSS2, Activates PAR-2 in Prostate Cancer Cells. Biochem. J. 2005, 388, 967–972. [Google Scholar] [CrossRef] [Green Version]
- Ko, C.J.; Huang, C.C.; Lin, H.Y.; Juan, C.P.; Lan, S.W.; Shyu, H.Y.; Wu, S.R.; Hsiao, P.W.; Huang, H.P.; Shun, C.T.; et al. Androgen-Induced TMPRSS2 Activates Matriptase and Promotes Extracellular Matrix Degradation, Prostate Cancer Cell Invasion, Tumor Growth, and Metastasis. Cancer Res. 2015, 75, 2949–2960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pohlmann, S. TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 2014, 88, 1293–1307. [Google Scholar] [CrossRef] [Green Version]
- Bertram, S.; Dijkman, R.; Habjan, M.; Heurich, A.; Gierer, S.; Glowacka, I.; Welsch, K.; Winkler, M.; Schneider, H.; Hofmann-Winkler, H.; et al. TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-Independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium. J. Virol. 2013, 87, 6150–6160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirato, K.; Kawase, M.; Matsuyama, S. Middle East Respiratory Syndrome Coronavirus Infection Mediated by the Transmembrane Serine Protease TMPRSS2. J. Virol. 2013, 87, 12552–12561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garten, W.; Braden, C.; Arendt, A.; Peitsch, C.; Baron, J.; Lu, Y.; Pawletko, K.; Hardes, K.; Steinmetzer, T.; Böttcher-Friebertshäuser, E. Influenza Virus Activating Host Proteases: Identification, Localization and Inhibitors as Potential Therapeutics. Eur. J. Cell Biol. 2015, 94, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Tahara, M.; Sakai, K.; Yamaguchi, H.; Kanou, K.; Shirato, K.; Kawase, M.; Noda, M.; Kimura, H.; Matsuyama, S.; et al. TMPRSS2 Is an Activating Protease for Respiratory Parainfluenza Viruses. J. Virol. 2013, 87, 11930–11935. [Google Scholar] [CrossRef] [Green Version]
- Vaarala, M.H.; Porvari, K.S.; Kellokumpu, S.; Kyllönen, A.P.; Vihko, P.T. Expression of Transmembrane Serine Protease TMPRSS2 in Mouse and Human Tissues. J. Pathol. 2001, 193, 134–140. [Google Scholar] [CrossRef]
- Chen, Y.W.; Lee, M.S.; Lucht, A.; Chou, F.P.; Huang, W.; Havighurst, T.C.; Kim, K.; Wang, J.K.; Antalis, T.M.; Johnson, M.D.; et al. TMPRSS2, a Serine Protease Expressed in the Prostate on the Apical Surface of Luminal Epithelial Cells and Released into Semen in Prostasomes, Is Misregulated in Prostate Cancer Cells. Am. J. Pathol. 2010, 176, 2986–2996. [Google Scholar] [CrossRef]
- Zang, R.; Castro, M.F.G.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 Promote SARS-CoV-2 Infection of Human Small Intestinal Enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef] [PubMed]
- Asselta, R.; Paraboschi, E.M.; Mantovani, A.; Duga, S. ACE2 and TMPRSS2 Variants and Expression as Candidates to Sex and Country Differences in COVID-19 Severity in Italy. Aging (Albany NY) 2020, 12, 10087–10098. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Blazyte, A.; Yoon, C.; Ryu, H.; Jeon, Y.; Bhak, Y.; Bolser, D.; Manica, A.; Shin, E.-S.; Cho, Y.S.; et al. Ethnicity-Dependent Allele Frequencies Are Correlated with COVID-19 Case Fatality Rate. Res. Sq. 2020, 1. [Google Scholar] [CrossRef]
- Paniri, A.; Hosseini, M.M.; Akhavan-Niaki, H. First Comprehensive Computational Analysis of Functional Consequences of TMPRSS2 SNPs in Susceptibility to SARS-CoV-2 among Different Populations. J. Biomol. Struct. Dyn. 2020, 1–18. [Google Scholar] [CrossRef]
- Senapati, S.; Kumar, S.; Singh, A.K.; Banerjee, P.; Bhagavatula, S. Assessment of Risk Conferred by Coding and Regulatory Variations of TMPRSS2 and CD26 in Susceptibility to SARS-CoV-2 Infection in Human. J. Genet. 2020, 99, 1–5. [Google Scholar] [CrossRef]
- Latini, A.; Agolini, E.; Novelli, A.; Borgiani, P.; Giannini, R.; Gravina, P.; Smarrazzo, A.; Dauri, M.; Andreoni, M.; Rogliani, P.; et al. COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells. Genes 2020, 11, 1010. [Google Scholar] [CrossRef]
- Al-Mulla, F.; Mohammad, A.; Al Madhoun, A.; Haddad, D.; Ali, H.; Eaaswarkhanth, M.; John, S.E.; Nizam, R.; Channanath, A.; Abu-Farha, M.; et al. ACE2 and FURIN Variants Are Potential Predictors of SARS-CoV-2 Outcome: A Time to Implement Precision Medicine against COVID-19. Heliyon 2021, 7, e06133. [Google Scholar] [CrossRef]
- Daga, S.; Fallerini, C.; Baldassarri, M.; Fava, F.; Valentino, F.; Doddato, G.; Benetti, E.; Furini, S.; Giliberti, A.; Tita, R.; et al. Employing a Systematic Approach to Biobanking and Analyzing Clinical and Genetic Data for Advancing COVID-19 Research. Eur. J. Human Genet. 2021, 1–15. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Herter, S.; Piper, D.E.; Aaron, W.; Gabriele, T.; Cutler, G.; Cao, P.; Bhatt, A.S.; Choe, Y.; Craik, C.S.; Walker, N.; et al. Hepatocyte Growth Factor Is a Preferred in Vitro Substrate for Human Hepsin, a Membrane-Anchored Serine Protease Implicated in Prostate and Ovarian Cancers. Biochem. J. 2005, 390, 125–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLano, W.L. The PyMOL Molecular Graphics System; Version 2.3; Schrödinger LLC: New York, NY, USA, 2020. [Google Scholar]
- Pandurangan, A.P.; Ochoa-Montaño, B.; Ascher, D.B.; Blundell, T.L. SDM: A Server for Predicting Effects of Mutations on Protein Stability. Nucleic Acids Res. 2017, 45, W229–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A Method and Server for Predicting Damaging Missense Mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Cimmaruta, C.; Citro, V.; Andreotti, G.; Liguori, L.; Cubellis, M.V.; Hay Mele, B. Challenging Popular Tools for the Annotation of Genetic Variations with a Real Case, Pathogenic Mutations of Lysosomal Alpha-Galactosidase. BMC Bioinf. 2018, 19, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Zmora, P.; Moldenhauer, A.S.; Hofmann-Winkler, H.; Pöhlmann, S. TMPRSS2 Isoform 1 Activates Respiratory Viruses and Is Expressed in Viral Target Cells. PLoS ONE 2015, 10, e0138380. [Google Scholar] [CrossRef] [Green Version]
- The Severe Covid-19 GWAS Group; Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernández, J.; Prati, D.; Baselli, G.; et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting Amino Acid Changes That Affect Protein Function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resnick, D.; Pearson, A.; Krieger, M. The SRCR Superfamily: A Family Reminiscent of the Ig Superfamily. Trends Biochem. Sci. 1994, 19, 5–8. [Google Scholar] [CrossRef]
- Hohenester, E.; Sasaki, T.; Timpl, R. Crystal Structure of a Scavenger Receptor Cysteine-Rich Domain Sheds Light on an Ancient Superfamily. Nat. Struct. Biol. 1999, 6, 228–232. [Google Scholar] [CrossRef]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.P.; George, N.; Fexova, S.; Fonseca, N.A.; Füllgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas Update: From Tissues to Single Cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, K.; Stoffel, W. TMbase: A Database of Membrane Spanning Protein Segments. Biol. Chem. 1993, 374, 166. [Google Scholar]
- Cattrini, C.; Bersanelli, M.; Latocca, M.M.; Conte, B.; Vallome, G.; Boccardo, F. Sex Hormones and Hormone Therapy during Covid-19 Pandemic: Implications for Patients with Cancer. Cancers 2020, 12, 2325. [Google Scholar] [CrossRef]
- Salciccia, S.; Del Giudice, F.; Eisenberg, M.L.; Mastroianni, C.M.; De Berardinis, E.; Ricciuti, G.P.; Maggi, M.; Sciarra, A. Androgen-Deprivation Therapy and SARS-Cov-2 Infection: The Potential Double-Face Role of Testosterone. Ther. Adv. Endocrinol. Metab. 2020, 11, 11. [Google Scholar] [CrossRef]
- Seeland, U.; Coluzzi, F.; Simmaco, M.; Mura, C.; Bourne, P.E.; Heiland, M.; Preissner, R.; Preissner, S. Evidence for Treatment with Estradiol for Women with SARS-CoV-2 Infection. BMC Med. 2020, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Breining, P.; Frølund, A.L.; Højen, J.F.; Gunst, J.D.; Staerke, N.B.; Saedder, E.; Cases-Thomas, M.; Little, P.; Nielsen, L.P.; Søgaard, O.S.; et al. Camostat Mesylate against SARS-CoV-2 and COVID-19—Rationale, Dosing and Safety. Basic Clin. Pharmacol. Toxicol. 2021, 128. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hofmann-Winkler, H.; Smith, J.C.; Krüger, N.; Sørensen, L.K.; Søgaard, O.S.; Hasselstrøm, J.B.; Winkler, M.; Hempel, T.; Raich, L.; et al. Camostat Mesylate Inhibits SARS-CoV-2 Activation by TMPRSS2-Related Proteases and Its Metabolite GBPA Exerts Antiviral Activity. bioRxiv 2020, 1. [Google Scholar] [CrossRef]
- Jang, S.; Rhee, J.Y. Three Cases of Treatment with Nafamostat in Elderly Patients with COVID-19 Pneumonia Who Need Oxygen Therapy. Int. J. Infect. Dis. 2020, 96, 500–502. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, W.; Yoneda, T.; Koba, H.; Ueda, T.; Tsuji, N.; Ogawa, H.; Asakura, H. Potential Mechanisms of Nafamostat Therapy for Severe COVID-19 Pneumonia with Disseminated Intravascular Coagulation. Int. J. Infect. Dis. 2021, 102, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Mele, B.H.; Citro, V.; Andreotti, G.; Cubellis, M.V. Drug Repositioning Can Accelerate Discovery of Pharmacological Chaperones. Orphanet J. Rare Dis. 2015, 10, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khadka, S.; Yuchi, A.; Shrestha, D.B.; Budhathoki, P.; Al-Subari, S.M.M.; Alhouzani, Z.T.M.; Butt, A.I. Repurposing Drugs for COVID-19: An Approach for Treatment in the Pandemic. Altern. Ther. Health Med. 2020, 26, 100–107. [Google Scholar]
- Tworowski, D.; Gorohovski, A.; Mukherjee, S.; Carmi, G.; Levy, E.; Detroja, R.; Mukherjee, S.B.; Frenkel-Morgenstern, M. COVID19 Drug Repository: Text-Mining the Literature in Search of Putative COVID19 Therapeutics. Nucleic Acids Res. 2021, 49, D1113–D1121. [Google Scholar] [CrossRef]
- Shrimp, J.H.; Kales, S.C.; Sanderson, P.E.; Simeonov, A.; Shen, M.; Hall, M.D. An Enzymatic TMPRSS2 Assay for Assessment of Clinical Candidates and Discovery of Inhibitors as Potential Treatment of COVID-19. ACS Pharmacol. Transl. Sci. 2020, 3, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Okajima, M.; Takahashi, Y.; Kaji, T.; Ogawa, N.; Mouri, H. Nafamostat Mesylate-Induced Hyperkalemia in Critically Ill Patients with COVID-19: Four Case Reports. World J. Clin. Cases 2020, 8, 5320–5325. [Google Scholar] [CrossRef]
- Singh, N.; Decroly, E.; Khatib, A.M.; Villoutreix, B.O. Structure-Based Drug Repositioning over the Human TMPRSS2 Protease Domain: Search for Chemical Probes Able to Repress SARS-CoV-2 Spike Protein Cleavages. Eur. J. Pharm. Sci. 2020, 153, 105495. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, I.; Russo, R.; Lasorsa, V.A.; Cantalupo, S.; Rosato, B.E.; Bonfiglio, F.; Frisso, G.; Abete, P.; Cassese, G.M.; Servillo, G.; et al. Common variants at 21q22.3 locus influence MX1 and TMPRSS2 gene expression and susceptibility to severe COVID-19. Iscience 2021, 102322. [Google Scholar] [CrossRef] [PubMed]


| (A) p.G296G | |||
|---|---|---|---|
| Mild | Severe | Marginal_Rows | |
| Wild-type | 306 (63%) | 490 (71%) | 796 |
| G296G | 178 (37%) | 198 (29%) | 376 |
| marginal_cols | 484 | 688 | 1172 |
| (B) p.V197M | |||
| Mild | Severe | Marginal_Rows | |
| Wild-type | 313 (64%) | 482 (70%) | 795 |
| V197M | 176 (36%) | 206 (30%) | 382 |
| marginal_cols | 489 | 688 | 1177 |
| (A) Male, Young (Age ≤ 60) | |||
|---|---|---|---|
| Mild | Severe | Marginal_Rows | |
| Wild-type | 102 (63%) | 140 (75%) | 242 |
| Val197Met | 59 (37%) | 47 (25%) | 106 |
| marginal_cols | 161 | 187 | 348 |
| (B) Female, Elderly (Age ≥ 58) | |||
| Mild | Severe | Marginal_Rows | |
| Wild-type | 37 (57%) | 131 (71%) | 168 |
| Val197Met | 28 (43%) | 53 (29%) | 81 |
| marginal_cols | 65 | 184 | 249 |
| (A) Male with Co-Morbidities, Young (Age ≤ 60) | |||
|---|---|---|---|
| Mild | Severe | Marginal_Rows | |
| Wild-type | 16 (50%) | 65 (77%) | 81 |
| Val197Met | 16 (50%) | 19 (23%) | 35 |
| marginal_cols | 32 | 84 | 116 |
| (B) Female with Co-Morbidities, Elderly (Age ≥ 58) | |||
| Mild | Severe | Marginal_Rows | |
| Wild-type | 21 (54%) | 105 (71%) | 126 |
| Val197Met | 18 (46%) | 42 (29%) | 60 |
| marginal_cols | 39 | 147 | 186 |
| (A) Male, Very Young (Age ≤ 50) | |||
|---|---|---|---|
| Mild | Severe | Marginal_Rows | |
| Wild-type | 7 (39%) | 12 (75%) | 19 |
| Val197Met | 11 (61%) | 4 (25%) | 15 |
| marginal_cols | 18 | 16 | 34 |
| (B) Female, Very Elderly (Age ≥ 71) | |||
| Mild | Severe | Marginal_Rows | |
| Wild-type | 6 (37.5%) | 66 (71%) | 72 |
| Val197Met | 10 (62.5%) | 27 (29%) | 37 |
| marginal_cols | 16 | 93 | 109 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monticelli, M.; Hay Mele, B.; Benetti, E.; Fallerini, C.; Baldassarri, M.; Furini, S.; Frullanti, E.; Mari, F.; GEN-COVID Multicenter Study; Andreotti, G.; et al. Protective Role of a TMPRSS2 Variant on Severe COVID-19 Outcome in Young Males and Elderly Women. Genes 2021, 12, 596. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12040596
Monticelli M, Hay Mele B, Benetti E, Fallerini C, Baldassarri M, Furini S, Frullanti E, Mari F, GEN-COVID Multicenter Study, Andreotti G, et al. Protective Role of a TMPRSS2 Variant on Severe COVID-19 Outcome in Young Males and Elderly Women. Genes. 2021; 12(4):596. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12040596
Chicago/Turabian StyleMonticelli, Maria, Bruno Hay Mele, Elisa Benetti, Chiara Fallerini, Margherita Baldassarri, Simone Furini, Elisa Frullanti, Francesca Mari, GEN-COVID Multicenter Study, Giuseppina Andreotti, and et al. 2021. "Protective Role of a TMPRSS2 Variant on Severe COVID-19 Outcome in Young Males and Elderly Women" Genes 12, no. 4: 596. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12040596