Heterozygous Nme7 Mutation Affects Glucose Tolerance in Male Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Genotyping
2.3. Experimental Protocol
2.3.1. Glucose Tolerance Tests
2.3.2. Biochemistry
2.3.3. Indirect Calorimetry
2.4. Gene Expression
2.5. Histology
2.6. Statistical Analysis
3. Results
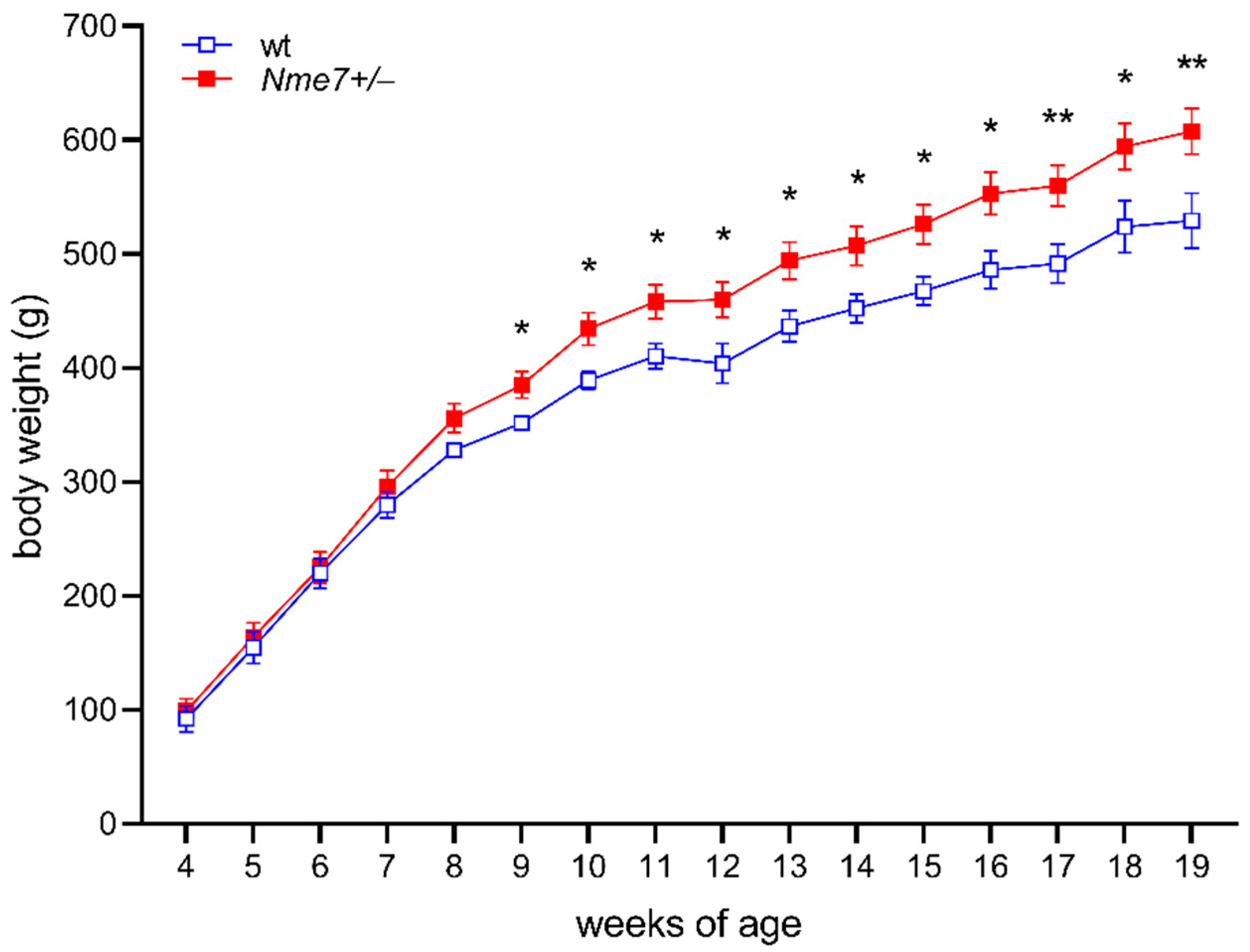
3.1. Increased Body Weight and Adiposity in Nme7+/− Rats
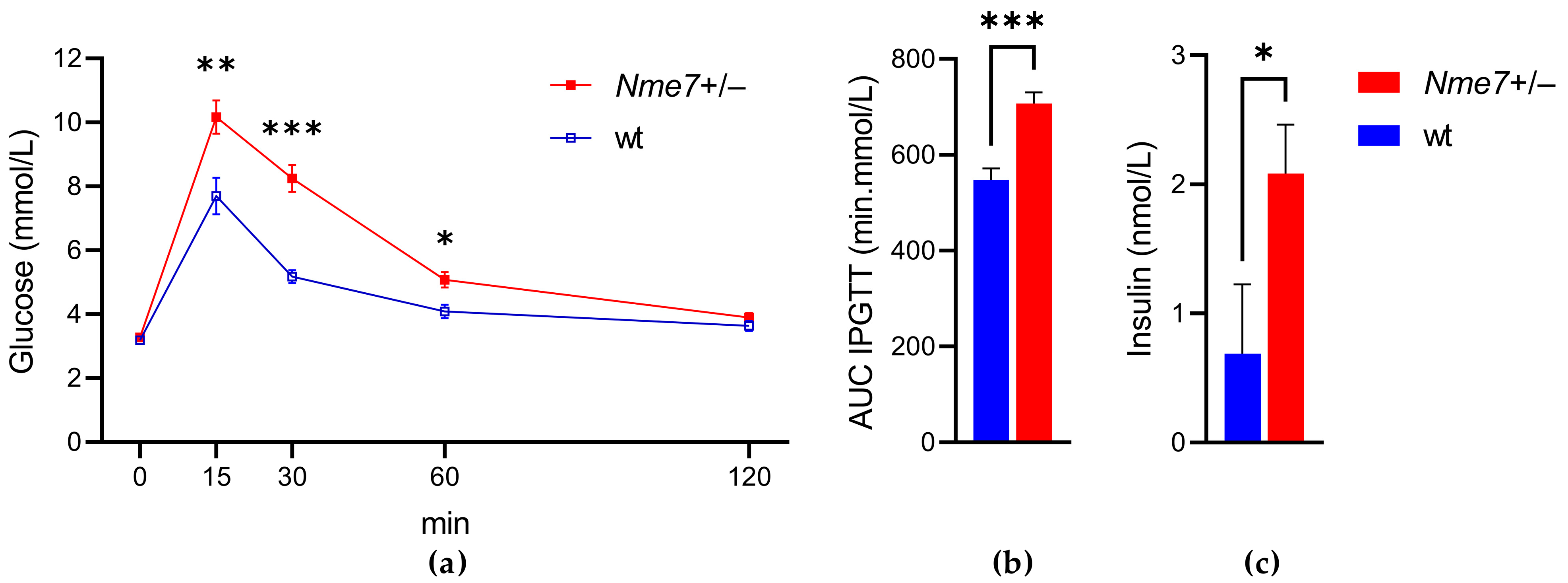
3.2. Impaired Glucose Tolerance of Nme7+/− Rats
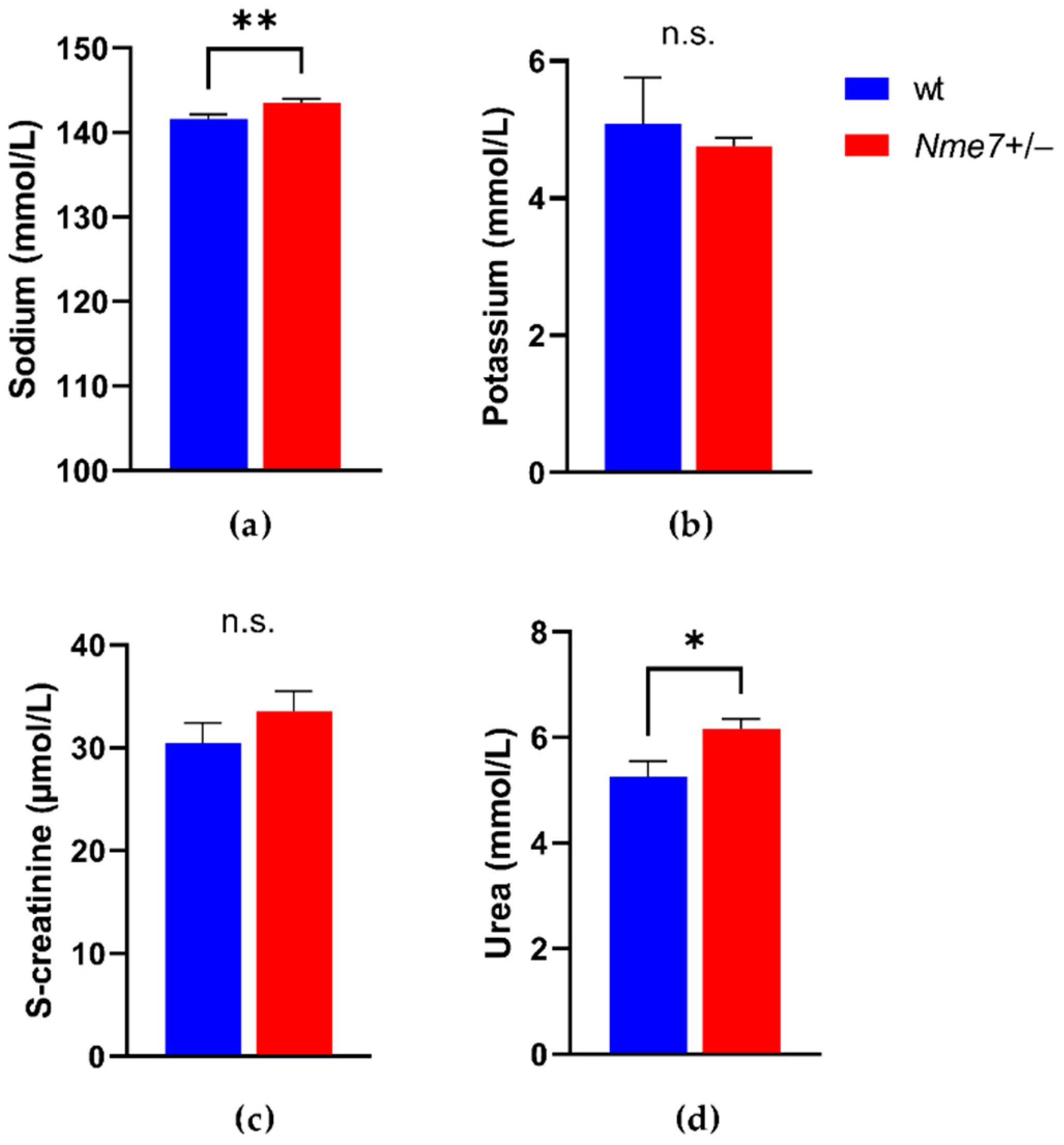
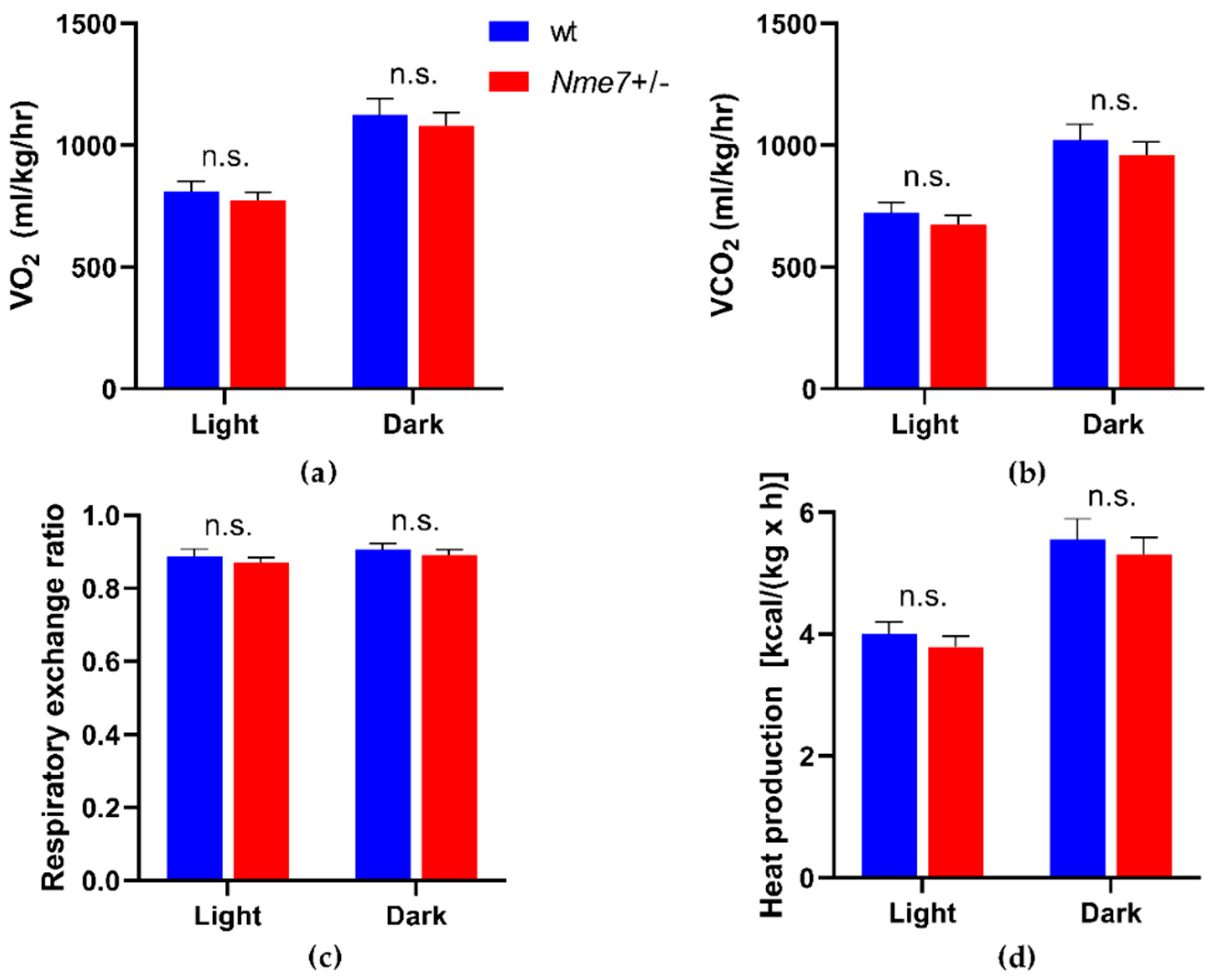
3.3. Metabolic and Biochemical Profile of Nme7+/− and wild-Type Rats
3.4. Histological Assessment of Nme7+/− and Wild-Type Male Rats
3.5. Expression of Nme7 and Transcriptome Profile of Nme7+/− and Wild-Type Rats
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blüher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Seda, O.; Liška, F.; Křenová, A.; Kazdová, L.; Šedová, L.; Zima, T.; Peng, J.; Pelinkova, K.; Tremblay, J.; Hamet, P.; et al. Dynamic genetic architecture of metabolic syndrome attributes in the rat. Physiol. Genom. 2005, 21, 243–252. [Google Scholar] [CrossRef] [Green Version]
- Kumar S., U.; Rajan, B.; Kumar D., T.; Preethi V., A.; Abunada, T.; Younes, S.; Okashah, S.; Ethiraj, S.; Priya Doss C., G.; Zayed, H. Involvement of Essential Signaling Cascades and Analysis of Gene Networks in Diabesity. Genes 2020, 11, 1256. [Google Scholar] [CrossRef]
- Nikpay, M.; Turner, A.W.; McPherson, R. Partitioning the Pleiotropy Between Coronary Artery Disease and Body Mass Index Reveals the Importance of Low Frequency Variants and Central Nervous System–Specific Functional Elements. Circ. Genom. Precis. Med. 2018, 11, e002050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Rasheed, A.; Tikkanen, E.; Lee, J.-J.; Butterworth, A.S.; Howson, J.M.M.; Assimes, T.L.; Chowdhury, R.; Orho-Melander, M.; Damrauer, S.; et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat. Genet. 2017, 49, 1450–1457. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; He, H.; Zhang, L.; Zhu, W.; Shen, H.; Yan, Y.-J.; Deng, H.-W. GWA-based pleiotropic analysis identified potential SNPs and genes related to type 2 diabetes and obesity. J. Hum. Genet. 2021, 66, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Birling, M.-C.; International Mouse Phenotyping Consortium (IMPC); Yoshiki, A.; Adams, D.J.; Ayabe, S.; Beaudet, A.L.; Bottomley, J.; Bradley, A.; Brown, S.D.M.; Bürger, A.; et al. A resource of targeted mutant mouse lines for 5061 genes. Nat. Genet. 2021, 53, 416–419. [Google Scholar] [CrossRef]
- Vcelak, J.; Seda, O.; Vankova, M.; Lukasova, P.; Vrbikova, J.; Tremblay, J.; Bendlova, B.; Hamet, P. Common variant on 1q24.2 (187cM) affects insulin secretion of beta cells and lipid spectrum in French-Canadian and Czech populations. Diabetologia 2009, 52, S230–S231. [Google Scholar]
- Hodulova, M.; Sedova, L.; Křenová, D.; Liška, F.; Krupkova, M.; Kazdová, L.; Tremblay, J.; Hamet, P.; Kren, V.; Šeda, O. Genomic Determinants of Triglyceride and Cholesterol Distribution into Lipoprotein Fractions in the Rat. PLoS ONE 2014, 9, e109983. [Google Scholar] [CrossRef] [PubMed]
- Šedová, L.; Školníková, E.; Hodúlová, M.; Včelák, J.; Šeda, O.; Bendlová, B. Expression Profiling of Nme7 Interactome in Experimental Models of Metabolic Syndrome. Physiol. Res. 2018, 67, S543–S550. [Google Scholar] [CrossRef]
- Ostrowski, L.E.; Blackburn, K.; Radde, K.M.; Moyer, M.B.; Schlatzer, D.M.; Moseley, A.; Boucher, R.C. A proteomic analysis of human cilia: Identification of novel components. Mol. Cell Proteom. 2002, 1, 451–465. [Google Scholar] [CrossRef] [Green Version]
- Lai, C.K.; Gupta, N.; Wen, X.; Rangell, L.; Chih, B.; Peterson, A.S.; Bazan, J.F.; Li, L.; Scales, S.J. Functional characterization of putative cilia genes by high-content analysis. Mol. Biol. Cell 2011, 22, 1104–1119. [Google Scholar] [CrossRef]
- Liu, P.; Choi, Y.K.; Qi, R.Z. NME7 is a functional component of the gamma-tubulin ring complex. Mol. Biol. Cell 2014, 25, 2017–2025. [Google Scholar] [CrossRef] [PubMed]
- Reish, O.; Aspit, L.; Zouella, A.; Roth, Y.; Polak-Charcon, S.; Baboushkin, T.; Benyamini, L.; Scheetz, T.; Mussaffi, H.; Sheffield, V.; et al. A Homozygous Nme7 Mutation Is Associated withSitus Inversus Totalis. Hum. Mutat. 2016, 37, 727–731. [Google Scholar] [CrossRef]
- Vogel, P.; Read, R.; Hansen, G.M.; Freay, L.C.; Zambrowicz, B.P.; Sands, A.T. Situs inversus in Dpcd/Poll-/-, Nme7-/-, and Pkd1l1-/- mice. Vet. Pathol. 2010, 47, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Vogel, P.; Read, R.W.; Hansen, G.M.; Payne, B.J.; Small, D.; Sands, A.T.; Zambrowicz, B.P. Congenital Hydrocephalus in Genetically Engineered Mice. Vet. Pathol. 2011, 49, 166–181. [Google Scholar] [CrossRef]
- Šedová, L.; Buková, I.; Bažantová, P.; Petrezsélyová, S.; Prochazka, J.; Školníková, E.; Zudová, D.; Včelák, J.; Makovický, P.; Bendlová, B.; et al. Semi-Lethal Primary Ciliary Dyskinesia in Rats Lacking the Nme7 Gene. Int. J. Mol. Sci. 2021, 22, 3810. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling The False Discovery Rate—A Practical And Powerful Approach To Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Boissan, M.; Schlattner, U.; Lacombe, M.-L. The NDPK/NME superfamily: State of the art. Lab. Investig. 2018, 98, 164–174. [Google Scholar] [CrossRef]
- Vujkovic, M.; Keaton, J.M.; Lynch, J.A.; Miller, D.R.; Zhou, J.; Tcheandjieu, C.; Huffman, J.E.; Assimes, T.L.; Lorenz, K.; Zhu, X.; et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020, 52, 680–691. [Google Scholar] [CrossRef]
- Veluthakal, R.; Kaetzel, D.; Kowluru, A. Nm23-H1 regulates glucose-stimulated insulin secretion in pancreatic beta-cells via Arf6-Rac1 signaling axis. Cell Physiol. Biochem. 2013, 32, 533–541. [Google Scholar] [CrossRef]
- Hoffmann, T.J.; Ehret, G.B.; Nandakumar, P.; Ranatunga, D.; Schaefer, C.; Kwok, P.-Y.; Iribarren, C.; Chakravarti, G.B.E.P.N.A.; Risch, D.R.C.S.C.I.N. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat. Genet. 2017, 49, 54–64. [Google Scholar] [CrossRef] [Green Version]
- Van Setten, J.; Verweij, N.; Mbarek, H.; Niemeijer, M.N.; Trompet, S.; Arking, D.E.; Brody, J.A.; Gandin, I.; Grarup, N.; Hall, L.M.; et al. Genome-wide association meta-analysis of 30,000 samples identifies seven novel loci for quantitative ECG traits. Eur. J. Hum. Genet. 2019, 27, 952–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heit, J.A.; Armasu, S.M.; Asmann, Y.W.; Cunningham, J.M.; Matsumoto, M.E.; Petterson, T.M.; De Andrade, M. A genome-wide association study of venous thromboembolism identifies risk variants in chromosomes 1q24.2 and 9q. J. Thromb. Haemost. 2012, 10, 1521–1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera-Rivero, M.; Stoll, M.; Hegenbarth, J.-C.; Rühle, F.; Limperger, V.; Junker, R.; Franke, A.; Hoffmann, P.; Shneyder, M.; Stach, M.; et al. Single- and Multimarker Genome-Wide Scans Evidence Novel Genetic Risk Modifiers for Venous Thromboembolism. Thromb. Haemost. 2021. [Google Scholar] [CrossRef]
- Hajdu, A.; Róna, G. Morphological Observations on Spontaneous Pancreatic Islet Changes in Rats. Diabetes 1967, 16, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, A.; Herr, F.; Rona, G. The functional significance of a spontaneous pancreatic islet change in aged rats. Diabetologia 1968, 4, 44–47. [Google Scholar] [CrossRef] [Green Version]
- Imaoka, M.; Jindo, T.; Takasaki, W. The Process and Development Mechanism of Age-related Fibrosis in the Pancreatic Islets of Sprague-Dawley Rats: Immunohistochemical Detection of Myofibroblasts and Suppression Effect by Estrogen Treatment. J. Toxicol. Pathol. 2013, 26, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Movassat, J.; Saulnier, C.; Serradas, P.; Portha, B. Impaired development of pancreatic beta-cell mass is a primary event during the progression to diabetes in the GK rat. Diabetologia 1997, 40, 916–925. [Google Scholar] [CrossRef] [Green Version]
- Ko, S.H.; Kwon, H.S.; Kim, S.R.; Moon, S.D.; Ahn, Y.B.; Song, K.H.; Son, H.S.; Cha, B.Y.; Lee, K.W.; Son, H.Y.; et al. Ramipril treatment suppresses islet fibrosis in Otsuka Long-Evans Tokushima fatty rats. Biochem. Biophys. Res. Commun. 2004, 316, 114–122. [Google Scholar] [CrossRef]
- Clark, A.; Nilsson, M.R. Islet amyloid: A complication of islet dysfunction or an aetiological factor in Type 2 diabetes? Diabetologia 2004, 47, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Kusaba, T.; Lalli, M.; Kramann, R.; Kobayashi, A.; Humphreys, B.D. Differentiated kidney epithelial cells repair injured proximal tubule. Proc. Natl. Acad. Sci. USA 2014, 111, 1527–1532. [Google Scholar] [CrossRef] [Green Version]
- Maeshima, A.; Yamashita, S.; Nojima, Y. Identification of renal progenitor-like tubular cells that participate in the regeneration processes of the kidney. J. Am. Soc. Nephrol. 2003, 14, 3138–3146. [Google Scholar] [CrossRef] [Green Version]
- Montserrat, N.; Ramirez-Bajo, M.J.; Xia, Y.; Sancho-Martinez, I.; Rull, D.M.; Serra, L.M.; Yang, S.; Nivet, E.; Cortina, C.; González, F.; et al. Generation of Induced Pluripotent Stem Cells from Human Renal Proximal Tubular Cells with Only Two Transcription Factors, Oct4 and Sox2. J. Biol. Chem. 2012, 287, 24131–24138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.-H.; Ma, N.; Lin, Y.-T.; Wu, C.-C.; Hsiao, M.; Lu, F.L.; Yu, C.-C.; Chen, S.-Y.; Lu, J. A shRNA Functional Screen Reveals Nme6 and Nme7 Are Crucial for Embryonic Stem Cell Renewal. STEM CELLS 2012, 30, 2199–2211. [Google Scholar] [CrossRef]
- Hinden, L.; Kogot-Levin, A.; Tam, J.; Leibowitz, G. Pathogenesis of diabesity-induced kidney disease: Role of kidney nutrient sensing. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kakiyama, G.; Marques, D.; Martin, R.; Takei, H.; Rodriguez-Agudo, D.; LaSalle, S.A.; Hashiguchi, T.; Liu, X.; Green, R.; Erickson, S.; et al. Insulin resistance dysregulates CYP7B1 leading to oxysterol accumulation: A pathway for NAFL to NASH transition. J. Lipid Res. 2020, 61, 1629–1644. [Google Scholar] [CrossRef]
- Powell, D.R.; Gay, J.P.; Smith, M.; Wilganowski, N.; Harris, A.; Holland, A.; Reyes, M.; Kirkham, L.; Kirkpatrick, L.L.; Zambrowicz, B.; et al. Fatty acid desaturase 1 knockout mice are lean with improved glycemic control and decreased development of atheromatous plaque. Diabetes Metab. Syndr. Obes. Targets Ther. 2016, 9, 185–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virbasius, J.; Czech, M.P. Map4k4 Signaling Nodes in Metabolic and Cardiovascular Diseases. Trends Endocrinol. Metab. 2016, 27, 484–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odom, D.T.; Zizlsperger, N.; Gordon, D.B.; Bell, G.W.; Rinaldi, N.J.; Murray, H.L.; Volkert, T.L.; Schreiber, J.; Rolfe, P.A.; Gifford, D.K.; et al. Control of pancreas and liver gene expression by HNF transcription factors. Science 2004, 303, 1378–1381. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šedová, L.; Prochazka, J.; Zudová, D.; Bendlová, B.; Včelák, J.; Sedlacek, R.; Šeda, O. Heterozygous Nme7 Mutation Affects Glucose Tolerance in Male Rats. Genes 2021, 12, 1087. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12071087
Šedová L, Prochazka J, Zudová D, Bendlová B, Včelák J, Sedlacek R, Šeda O. Heterozygous Nme7 Mutation Affects Glucose Tolerance in Male Rats. Genes. 2021; 12(7):1087. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12071087
Chicago/Turabian StyleŠedová, Lucie, Jan Prochazka, Dagmar Zudová, Běla Bendlová, Josef Včelák, Radislav Sedlacek, and Ondřej Šeda. 2021. "Heterozygous Nme7 Mutation Affects Glucose Tolerance in Male Rats" Genes 12, no. 7: 1087. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12071087





