Molecular Pathways Involved in the Development of Congenital Erythrocytosis
Abstract
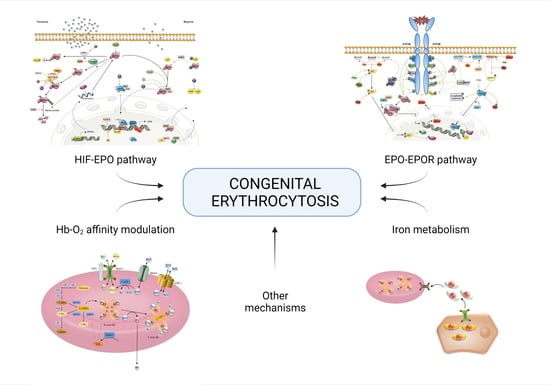
:1. Introduction
2. Known Molecular Pathways Involved in Congenital Erythrocytosis
2.1. Oxygen-Sensing: HIF-EPO Pathway
2.1.1. Regulation at the RNA Level
2.1.2. Regulation at the Protein Level
Protein Interactions and Modifications
Nuclear Transport
2.2. Signal Transduction: EPO-EPOR Pathway
2.2.1. Regulation at the RNA Level
2.2.2. Regulation at the Protein Level
Protein Interactions and Modifications in STAT5 Pathway
STAT5 Negative Regulation
Protein Interactions and Modifications in PI3K Pathway
PI3K/AKT Negative Regulation
Protein Interactions and Modifications in MAPK Pathway
MAPK Negative Regulation
2.3. Hemoglobin-Oxygen Affinity Modulation
2.3.1. Regulation at the RNA Level
2.3.2. Regulation at the Protein Level
Hemoglobin Structure and Function Modulation
Gas Transport
2.3.3. Regulation at the Metabolic Level: Glycolysis and Oxygen Affinity Regulators
3. Other Molecular Pathways Involved in Congenital Erythrocytosis
3.1. Regulation of Red Blood Cells Production and Degradation
3.2. Iron Metabolism
3.3. Other Mechanisms
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bento, C. Genetic Basis of Congenital Erythrocytosis. Int. J. Lab. Hematol. 2018, 40, 62–67. [Google Scholar] [CrossRef] [Green Version]
- Gašperšič, J.; Kristan, A.; Kunej, T.; Preložnik Zupan, I.; Debeljak, N. Erythrocytosis: Genes and Pathways Involved in Disease Development. Blood Transfus. 2020. [Google Scholar] [CrossRef]
- McMullin, M.F. Genetic Background of Congenital Erythrocytosis. Genes 2021, 12, 1151. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Iny Stein, T.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a Knowledge-Based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- NCBI, R.C. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef] [Green Version]
- Tweedie, S.; Braschi, B.; Gray, K.; Jones, T.E.M.; Seal, R.L.; Yates, B.; Bruford, E.A. Genenames.Org: The HGNC and VGNC Resources in 2021. Nucleic Acids Res. 2021, 49, D939–D946. [Google Scholar] [CrossRef] [PubMed]
- Beutler, E.; Westwood, B.; Van Zwieten, R.; Roos, D. G→t Transition at Cdna Nt 110 (K37q) in the Pklr (Pyruvate Kinase) Gene Is the Molecular Basis of a Case of Hereditary Increase of Red Blood Cell Atp. Hum. Mutat. 1997, 9, 282–285. [Google Scholar] [CrossRef]
- Biagetti, G.; Catherwood, M.; Robson, N.; Bertozzi, I.; Cosi, E.; McMullin, M.F.; Randi, M.L. HFE Mutations in Idiopathic Erythrocytosis. Br. J. Haematol. 2018, 181, 270–272. [Google Scholar] [CrossRef] [Green Version]
- Lappin, T.R.; Lee, F.S. Update on Mutations in the HIF: EPO Pathway and Their Role in Erythrocytosis. Blood Rev. 2019, 37, 100590. [Google Scholar] [CrossRef]
- Camps, C.; Petousi, N.; Bento, C.; Cario, H.; Copley, R.R.; McMullin, M.F.; Van Wijk, R.; Ratcliffe, P.J.; Robbins, P.A.; Taylor, J.C. Gene Panel Sequencing Improves the Diagnostic Work-up of Patients with Idiopathic Erythrocytosis and Identifies New Mutations. Haematologica 2016, 101, 1306–1318. [Google Scholar] [CrossRef]
- Zmajkovic, J.; Lundberg, P.; Nienhold, R.; Torgersen, M.L.; Sundan, A.; Waage, A.; Skoda, R.C. A Gain-of-Function Mutation in EPO in Familial Erythrocytosis. N. Engl. J. Med. 2018, 378, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Lenglet, M.; Robriquet, F.; Schwarz, K.; Camps, C.; Couturier, A.; Hoogewijs, D.; Buffet, A.; Knight, S.; Gad, S.; Couvé, S.; et al. Identification of a new VHL exon and complex splicing alterations in familial erythrocytosis or von Hippel-Lindau disease. Blood 2018, 132, 469–483. [Google Scholar] [CrossRef] [Green Version]
- Tomc, J.; Debeljak, N. Molecular Insights into the Oxygen-Sensing Pathway and Erythropoietin. Int. J. Mol. Sci. 2021, 22, 7074. [Google Scholar] [CrossRef] [PubMed]
- Laureau, M.O. SMART Servier Medical Art. Les Laboratories Servier. Available online: https://smart.servier.com (accessed on 16 July 2021).
- Moniz, S.; Bandarra, D.; Biddlestone, J.; Campbell, K.J.; Komander, D.; Bremm, A.; Rocha, S. Cezanne Regulates E2F1-Dependent HIF2α Expression. J. Cell Sci. 2015, 128, 3082–3093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wada, T.; Shimba, S.; Tezuka, M. Transcriptional Regulation of the Hypoxia Inducible Factor-2α (HIF-2α) Gene during Adipose Differentiation in 3T3-L1 Cells. Biol. Pharm. Bull. 2006, 29, 49–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ligęza, J.; Marona, P.; Gach, N.; Lipert, B.; Miekus, K.; Wilk, W.; Jaszczyński, J.; Stelmach, A.; Loboda, A.; Dulak, J.; et al. MCPIP1 Contributes to Clear Cell Renal Cell Carcinomas Development. Angiogenesis 2017, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.H.; Bao, Y.; Wang, X.; Yan, F.; Guo, S.; Ma, Y.; Xu, D.; Jin, L.; Xu, J.; Wang, J. Hypoxic-Stabilized EPAS1 Proteins Transactivate DNMT1 and Cause Promoter Hypermethylation and Transcription Inhibition of EPAS1 in Non-Small Cell Lung Cancer. FASEB J. 2018, 32, 6694–6705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metzen, E.; Stiehl, D.P.; Doege, K.; Marxsen, J.H.; Hellwig-Bürgel, T.; Jelkmann, W. Regulation of the Prolyl Hydroxylase Domain Protein 2 (Phd2/Egln-1) Gene: Identification of a Functional Hypoxia-Responsive Element. Biochem. J. 2005, 387, 711–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wielockx, B.; Meneses, A. PHD2: From Hypoxia Regulation to Disease Progression. Hypoxia 2016, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, S.; Charbonneau, M.; Grandmont, S.; Richard, D.E.; Dubois, C.M. Transforming Growth Factor Β1 Induces Hypoxia-Inducible Factor-1 Stabilization through Selective Inhibition of PHD2 Expression. J. Biol. Chem. 2006, 281, 24171–24181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zatyka, M.; Morrissey, C.; Kuzmin, I.; Lerman, M.I.; Latif, F.; Richards, F.M.; Maher, E.R. Genetic and Functional Analysis of the von Hippel-Lindau (VHL) Tumour Suppressor Gene Promoter. J. Med. Genet. 2002, 39, 463–472. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Cai, X.; Hu, B.; Mei, Z.; Zhang, D.; Ouyang, G.; Wang, J.; Zhang, W.; Xiao, W. Forkhead Transcription Factor 3a (FOXO3a) Modulates Hypoxia Signaling via up-Regulation of the von Hippel-Lindau Gene (VHL). J. Biol. Chem. 2016, 291, 25692–25705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Łuczak, M.W.; Roszak, A.; Pawlik, P.; Kȩdzia, H.; Lianeri, M.; Jagodziński, P.P. Increased Expression of HIF-1A and Its Implication in the Hypoxia Pathway in Primary Advanced Uterine Cervical Carcinoma. Oncol. Rep. 2011, 26, 1259–1264. [Google Scholar] [CrossRef] [PubMed]
- Galson, D.L.; Tsuchiya, T.; Tendler, D.S.; Huang, L.E.; Ren, Y.; Ogura, T.; Bunn, H.F. The Orphan Receptor Hepatic Nuclear Factor 4 Functions as a Transcriptional Activator for Tissue-Specific and Hypoxia-Specific Erythropoietin Gene Expression and Is Antagonized by EAR3/COUP-TF1. Mol. Cell. Biol. 1995, 15, 2135–2144. [Google Scholar] [CrossRef] [Green Version]
- Makita, T.; Hernandez-Hoyos, G.; Chen, T.H.P.; Wu, H.; Rothenberg, E.V.; Sucov, H.M. A Developmental Transition in Definitive Erythropoiesis: Erythropoietin Expression Is Sequentially Regulated by Retinoic Acid Receptors and HNF4. Genes Dev. 2001, 15, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Zhang, R.; Wu, X.; Hankinson, O. Roles of Coactivators in Hypoxic Induction of the Erythropoietin Gene. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [Green Version]
- Imagawa, S.; Yamamoto, M.; Miura, Y. Negative Regulation of the Erythropoietin Gene Expression by the GATA Transcription Factors. Blood 1997, 89, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- Obara, N.; Suzuki, N.; Kim, K.; Nagasawa, T.; Imagawa, S.; Yamamoto, M. Repression via the GATA Box Is Essential for Tissue-Specific Erythropoietin Gene Expression. Blood 2008, 111, 5223–5232. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Blachard, K.L. Erratum: DNA Methylation Represses the Expression of the Human Erythropoietin Gene by Two Different Mechanisms. Blood 2000, 95, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Dengler, V.L.; Galbraith, M.; Espinosa, J.M. Transcriptional Regulation by Hypoxia Inducible Factors. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Gradin, K.; Takasaki, C.; Fujii-kuriyama, Y.; Sogawa, K. The Transcriptional Activation Function of the HIF-like Factor Requires Phosphorylation at a Conserved Threonine. J. Biol. Chem. 2002, 277, 23508–23514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pangou, E.; Befani, C.; Mylonis, I.; Samiotaki, M.; Panayotou, G.; Simos, G.; Liakos, P. HIF-2α Phosphorylation by CK1δ Promotes Erythropoietin Secretion in Liver Cancer Cells under Hypoxia. J. Cell Sci. 2016, 129, 4213–4226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dioum, E.M.; Chen, R.; Alexander, M.S.; Zhang, Q.; Hogg, R.T.; Gerard, R.D.; Garcia, J.A. Regulation of Hypoxia-Inducible Factor 2α Signaling by the Stress-Responsive Deacetylase Sirtuin 1. Science 2009, 324, 1289–1293. [Google Scholar] [CrossRef]
- Bouras, T.; Fu, M.; Sauve, A.A.; Wang, F.; Quong, A.A.; Perkins, N.D.; Hay, R.T.; Gu, W.; Pestell, R.G. SIRT1 Deacetylation and Repression of P300 Involves Lysine Residues 1020/1024 within the Cell Cycle Regulatory Domain 1. J. Biol. Chem. 2005, 280, 10264–10276. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Zhu, M.; Song, K.; Wuren, T.; Yan, J.; Ge, R.-L.; Ji, L.; Cui, S. VHL Gene Methylation Contributes to Excessive Erythrocytosis in Chronic Mountain Sickness Rat Model by Upregulating the HIF-2α/EPO Pathway. Life Sci. 2021, 266, 118873. [Google Scholar] [CrossRef]
- Kubaichuk, K.; Kietzmann, T. Involvement of E3 Ligases and Deubiquitinases in the Control of HIF-α Subunit Abundance. Cells 2019, 8, 598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Hagen, M.; Overmeer, R.M.; Abolvardi, S.S.; Vertegaal, A.C.O. RNF4 and VHL Regulate the Proteasomal Degradation of SUMO-Conjugated Hypoxia-Inducible Factor-2α. Nucleic Acids Res. 2009, 38, 1922–1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Han, Y.; Wang, Y.; Sun, X.; Yan, S.; Yeh, E.T.H.; Chen, Y.; Cang, H.; Li, H.; Shi, G.; et al. SENP3 Is Responsible for HIF-1 Transactivation under Mild Oxidative Stress via P300 de-SUMOylation. EMBO J. 2009, 28, 2748–2762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisy, K.; Peet, D.J. Turn Me on: Regulating HIF Transcriptional Activity. Cell Death Differ. 2008, 15, 642–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montagner, M.; Enzo, E.; Forcato, M.; Zanconato, F.; Parenti, A.; Rampazzo, E.; Basso, G.; Leo, G.; Rosato, A.; Bicciato, S.; et al. SHARP1 Suppresses Breast Cancer Metastasis by Promoting Degradation of Hypoxia-Inducible Factors. Nature 2012, 487, 380–384. [Google Scholar] [CrossRef]
- Barth, S.; Nesper, J.; Hasgall, P.A.; Wirthner, R.; Nytko, K.J.; Edlich, F.; Katschinski, D.M.; Stiehl, D.P.; Wenger, R.H.; Camenisch, G. The Peptidyl Prolyl Cis/Trans Isomerase FKBP38 Determines Hypoxia-Inducible Transcription Factor Prolyl-4-Hydroxylase PHD2 Protein Stability. Mol. Cell. Biol. 2007, 27, 3758–3768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, J.H.; Mahon, P.C.; Oh, J.; Kelly, B.; Krishnamachary, B.; Pearson, M.; Chan, D.A.; Giaccia, A.J.; Semenza, G.L. OS-9 Interacts with Hypoxia-Inducible Factor 1α and Prolyl Hydroxylases to Promote Oxygen-Dependent Degradation of HIF-1α. Mol. Cell 2005, 17, 503–512. [Google Scholar] [CrossRef]
- Kumar, P.; Gullberg, U.; Olsson, I.; Ajore, R. Myeloid Translocation Gene-16 Co-Repressor Promotes Degradation of Hypoxia-Inducible Factor 1. PLoS ONE 2015, 10, e0123725. [Google Scholar] [CrossRef] [Green Version]
- Minervini, G.; Quaglia, F.; Tosatto, S.C.E. Computational Analysis of Prolyl Hydroxylase Domain-Containing Protein 2 (PHD2) Mutations Promoting Polycythemia Insurgence in Humans. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Baek, J.H.; Liu, Y.V.; McDonald, K.R.; Wesley, J.B.; Hubbi, M.E.; Byun, H.; Semenza, G.L. Spermidine/Spermine-N1-Acetyltransferase 2 Is an Essential Component of the Ubiquitin Ligase Complex That Regulates Hypoxia-Inducible Factor 1alpha. J. Biol. Chem. 2007, 282, 23572–23580. [Google Scholar] [CrossRef] [Green Version]
- Gossage, L.; Eisen, T.; Maher, E.R. VHL, the Story of a Tumour Suppressor Gene. Nat. Rev. Cancer 2015, 15, 55–64. [Google Scholar] [CrossRef]
- Cardote, T.A.F.; Gadd, M.S.; Ciulli, A. Crystal Structure of the Cul2-Rbx1-EloBC-VHL Ubiquitin Ligase Complex. Structure 2017, 25, 901–911.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partch, C.L.; Gardner, K.H. Coactivators Necessary for Transcriptional Output of the Hypoxia Inducible Factor, HIF, Are Directly Recruited by ARNT PAS-B. Proc. Natl. Acad. Sci. USA 2011, 108, 7739–7744. [Google Scholar] [CrossRef] [Green Version]
- Depping, R.; Jelkmann, W.; Kosyna, F.K. Nuclear-Cytoplasmatic Shuttling of Proteins in Control of Cellular Oxygen Sensing. J. Mol. Med. 2015, 93, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Depping, R.; Steinhoff, A.; Schindler, S.G.; Friedrich, B.; Fagerlund, R.; Metzen, E.; Hartmann, E.; Köhler, M. Nuclear Translocation of Hypoxia-Inducible Factors (HIFs): Involvement of the Classical Importin α/β Pathway. Biochim. Biophys. Acta Mol. Cell Res. 2008, 1783, 394–404. [Google Scholar] [CrossRef] [Green Version]
- Gkotinakou, I.M.; Befani, C.; Simos, G.; Liakos, P. ERK1/2 Phosphorylates HIF-2α and Regulates Its Activity by Controlling Its CRM1-Dependent Nuclear Shuttling. J. Cell Sci. 2019, 132, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khacho, M.; Mekhail, K.; Pilon-Larose, K.; Pause, A.; Cote, J.; Lee, S. EEF1A Is a Novel Component of the Mammalian Nuclear Protein Export Machinery. Mol. Biol. Cell 2008, 19, 5296–5308. [Google Scholar] [CrossRef] [Green Version]
- Chin, H.; Arai, A.; Wakao, H.; Kamiyama, R.; Miyasaka, N.; Miura, O. Lyn Physically Associates with the Erythropoietin Receptor and May Play a Role in Activation of the Stat5 Pathway. Blood 1998, 91, 3734–3745. [Google Scholar] [CrossRef]
- Laubach, J.P.; Fu, P.; Jiang, X.; Salter, K.H.; Potti, A.; Arcasoy, M.O. Polycythemia Vera Erythroid Precursors Exhibit Increased Proliferation and Apoptosis Resistance Associated with Abnormal RAS and PI3K Pathway Activation. Exp. Hematol. 2009, 37, 1411–1422. [Google Scholar] [CrossRef] [Green Version]
- Debeljak, N.; Solár, P.; Sytkowski, A.J. Erythropoietin and Cancer: The Unintended Consequences of Anemia Correction. Front. Immunol. 2014, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Tóthová, Z.; Tomc, J.; Debeljak, N.; Solar, P. STAT5 as a Key Protein of Erythropoietin Signalization. Int. J. Mol. Sci. 2021, 22, 7109. [Google Scholar] [CrossRef]
- Tóthová, Z.; Šemeláková, M.; Solárová, Z.; Tomc, J.; Debeljak, N.; Solár, P. The Role of PI3K/AKT and MAPK Signaling Pathways in Erythropoietin Signalization. Int. J. Mol. Sci. 2021, 22, 7682. [Google Scholar] [CrossRef] [PubMed]
- Vočanec, D.; Prijatelj, T.; Debeljak, N.; Kunej, T. Genetic Variants of Erythropoietin (EPO) and EPO Receptor Genes in Familial Erythrocytosis. Int. J. Lab. Hematol. 2019, 41, 162–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodges, V.M.; Rainey, S.; Lappin, T.R.; Maxwell, A.P. Pathophysiology of Anemia and Erythrocytosis. Crit. Rev. Oncol. Hematol. 2007, 64, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Spolverini, A.; Pieri, L.; Guglielmelli, P.; Pancrazzi, A.; Fanelli, T.; Paoli, C.; Bosi, A.; Nichele, I.; Ruggeri, M.; Vannucchi, A.M. Infrequent Occurrence of Mutations in the PH Domain of LNK in Patients with JAK2 Mutation-Negative “idiopathic” Erythrocytosis. Haematologica 2013, 98, 101–102. [Google Scholar] [CrossRef] [PubMed]
- McMullin, M.F.; Cario, H. LNK mutations and myeloproliferative disorders. Am. J. Hematol. 2016, 91, 248–251. [Google Scholar] [CrossRef]
- Jegalian, A.G.; Wu, H. Differential Roles of SOCS Family Members in EpoR Signal Transduction. J. Interf. Cytokine Res. 2002, 22, 853–860. [Google Scholar] [CrossRef]
- Kapralova, K.; Horvathova, M.; Pecquet, C.; Fialova Kucerova, J.; Pospisilova, D.; Leroy, E.; Kralova, B.; Milosevic Feenstra, J.D.; Schischlik, F.; Kralovics, R.; et al. Cooperation of germ line JAK2 mutations E846D and R1063H in hereditary erythrocytosis with megakaryocytic atypia. Blood 2016, 128, 1418–1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuckey, R.; Gómez-Casares, T.G. Recent Advances in the Use of Molecular Analyses to Inform the Diagnosis and Prognosis of Patients with Polycythaemia Vera. Int. J. Mol. Sci. 2021, 22, 5042. [Google Scholar] [CrossRef]
- Merkle, R.; Steiert, B.; Salopiata, F.; Depner, S.; Raue, A.; Iwamoto, N.; Schelker, M.; Hass, H.; Wäsch, M.; Böhm, M.E.; et al. Identification of Cell Type-Specific Differences in Erythropoietin Receptor Signaling in Primary Erythroid and Lung Cancer Cells. PLoS Comput. Biol. 2016, 12, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Zon, L.I.; Youssoufian, H.; Mather, C.; Lodish, H.F.; Orkin, S.H. Activation of the Erythropoietin Receptor Promoter by Transcription Factor GATA-1. Proc. Natl. Acad. Sci. USA 1991, 88, 10638–10641. [Google Scholar] [CrossRef] [Green Version]
- Chin, K.; Oda, N.; Shen, K.; Tom noguchi, C. Regulation of Transcription of the Human Erythropoietin Receptor Gene by Proteins Binding to GATA-1 and Sp1 Motifs. Nucleic Acids Res. 1995, 23, 3041–3049. [Google Scholar] [CrossRef] [Green Version]
- Kuhrt, D.; Wojchowski, D.M. Emerging EPO and EPO Receptor Regulators and Signal Transducers. Blood 2015, 125, 3536–3541. [Google Scholar] [CrossRef] [Green Version]
- Perreault, A.A.; Benton, M.L.; Koury, M.J.; Brandt, S.J.; Venters, B.J. Epo Reprograms the Epigenome of Erythroid Cells. Exp. Hematol. 2017, 51, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Perreaulta, A.A.; Venters, B.J. Integrative View on How Erythropoietin Signaling Controls Transcription Patterns in Erythroid Cells. Curr. Opin. Hematol. 2018, 25, 189–195. [Google Scholar] [CrossRef]
- Maurer, B.; Kollmann, S.; Pickem, J.; Hoelbl-Kovacic, A.; Sexl, V. STAT5A and STAT5B—Twins with Different Personalities in Hematopoiesis and Leukemia. Cancers (Basel) 2019, 11, 1726. [Google Scholar] [CrossRef] [Green Version]
- Watowich, S.S. The Erythropoietin Receptor: Molecular Structure and Hematopoietic Signaling Pathways. J. Investig. Med. 2011, 59, 1067–1072. [Google Scholar] [CrossRef]
- Able, A.A.; Burrell, J.A.; Stephens, J.M. STAT5-Interacting Proteins: A Synopsis of Proteins That Regulate STAT5 Activity. Biology (Basel) 2017, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Ernst, S.; Müller-Newen, G. Nucleocytoplasmic Shuttling of Stats. A Target for Intervention? Cancers (Basel) 2019, 11, 1815. [Google Scholar] [CrossRef] [Green Version]
- Gillinder, K.R.; Tuckey, H.; Bell, C.C.; Magor, G.W.; Huang, S.; Ilsley, M.D.; Perkins, A.C. Direct Targets of PStat5 Signalling in Erythropoiesis. PLoS ONE 2017, 12, e0180922. [Google Scholar] [CrossRef] [PubMed]
- Okutani, Y.; Kitanaka, A.; Tanaka, T.; Kamano, H.; Ohnishi, H.; Kubota, Y.; Ishida, T.; Takahara, J. Src Directly Tyrosine-Phosphorylates STAT5 on Its Activation Site and Is Involved in Erythropoietin-Induced Signaling Pathway. Oncogene 2001, 20, 6643–6650. [Google Scholar] [CrossRef] [Green Version]
- Nagao, T.; Kurosu, T.; Umezawa, Y.; Nogami, A.; Oshikawa, G.; Tohda, S.; Yamamoto, M.; Miura, O. Proliferation and Survival Signaling from Both Jak2-V617F and Lyn Involving GSK3 and MTOR/P70S6K/4EBP1 in PVTL-1 Cell Line Newly Established from Acute Myeloid Leukemia Transformed from Polycythemia Vera. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ota, J.; Kimura, F.; Sato, K.; Wakimoto, N.; Nakamura, Y.; Nagata, N.; Suzu, S.; Yamada, M.; Shimamura, S.; Motoyoshi, K. Association of CrkL with STAT5 in Hematopoietic Cells Stimulated by Granulocyte-Macrophage Colony-Stimulating Factor or Erythropoietin. Biochem. Biophys. Res. Commun. 1998, 252, 779–786. [Google Scholar] [CrossRef]
- Held, M.A.; Greenfest-allen, E.; Su, S.; Stoeckert, C.J.; Stokes, M.P.; Wojchowski, D.M. Phospho-PTM Proteomic Discovery of Novel EPO- Modulated Kinases and Phosphatases, Including PTPN18 as a Positive Regulator of EPOR/JAK2 Signaling. Cell. Signal. 2020, 69, 109554. [Google Scholar] [CrossRef]
- Tong, W.; Zhang, J.; Lodish, H.F. Lnk Inhibits Erythropoiesis and Epo-Dependent JAK2 Activation and Downstream Signaling Pathways. Blood 2005, 105, 4604–4612. [Google Scholar] [CrossRef] [PubMed]
- Klingmüller, U.; Lorenz, U.; Cantley, L.C.; Neel, B.G.; Lodish, H.F. Specific Recruitment of SH-PTP1 to the Erythropoietin Receptor Causes Inactivation of JAK2 and Termination of Proliferative Signals. Cell 1995, 80, 729–738. [Google Scholar] [CrossRef] [Green Version]
- Ingley, E. Integrating Novel Signaling Pathways Involved in Erythropoiesis. IUBMB Life 2012, 64, 402–410. [Google Scholar] [CrossRef]
- Wojchowski, D.M.; Sathyanarayana, P.; Dev, A. Erythropoietin Receptor Response Circuits. Curr. Opin. Hematol. 2010, 17, 169–176. [Google Scholar] [CrossRef]
- Peltola, K.J.; Paukku, K.; Aho, T.L.T.; Ruuska, M.; Silvennoinen, O.; Koskinen, P.J. Pim-1 Kinase Inhibits STAT5-Dependent Transcription via Its Interactions with SOCS1 and SOCS3. Blood 2004, 103, 3744–3750. [Google Scholar] [CrossRef]
- Xie, Y.; Shi, X.; Sheng, K.; Han, G.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt Signaling Transduction Pathway, Erythropoiesis and Glycolysis in Hypoxia (Review). Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef] [Green Version]
- Bouscary, D.; Pene, F.; Claessens, Y.E.; Muller, O.; Chrétien, S.; Fontenay-Roupie, M.; Gisselbrecht, S.; Mayeux, P.; Lacombe, C. Critical Role for PI 3-Kinase in the Control of Erythropoietin-Induced Erythroid Progenitor Proliferation. Blood 2003, 101, 3436–3443. [Google Scholar] [CrossRef] [PubMed]
- Jafari, M.; Ghadami, E.; Dadkhah, T.; Akhavan-Niaki, H. PI3k/AKT Signaling Pathway: Erythropoiesis and Beyond. J. Cell. Physiol. 2019, 234, 2373–2385. [Google Scholar] [CrossRef]
- Verdier, F.; Chrétien, S.; Billat, C.; Gisselbrecht, S.; Lacombe, C.; Mayeux, P. Erythropoietin Induces the Tyrosine Phosphorylation of Insulin Receptor Substrate-2. An Alternate Pathway for Erythropoietin-Induced Phosphatidylinositol 3-Kinase Activation. J. Biol. Chem. 1997, 272, 26173–26178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Missiroli, S.; Etro, D.; Buontempo, F.; Ye, K.; Capitani, S.; Neri, L.M. Nuclear Translocation of Active AKT Is Required for Erythroid Differentiation in Erythropoietin Treated K562 Erythroleukemia Cells. Int. J. Biochem. Cell Biol. 2009, 41, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Leslie, N.R.; Biondi, R.M.; Alessi, D.R. Phosphoinositide-Regulated Kinases and Phosphoinositide Phosphatases. ChemInform 2010, 32. [Google Scholar] [CrossRef]
- Maira, S.M.; Galetic, I.; Brazil, D.P.; Kaech, S.; Ingley, E.; Thelen, M.; Hemmings, B.A. Carboxyl-Terminal Modulator Protein (CTMP), a Negative Regulator of PKB/Akt and v-Akt at the Plasma Membrane. Science 2001, 294, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Karayel, Ö.; Xu, P.; Bludau, I.; Velan Bhoopalan, S.; Yao, Y.; Ana Rita, F.C.; Santos, A.; Schulman, B.A.; Alpi, A.F.; Weiss, M.J.; et al. Integrative Proteomics Reveals Principles of Dynamic Phosphosignaling Networks in Human Erythropoiesis. Mol. Syst. Biol. 2020, 16, 1–22. [Google Scholar] [CrossRef]
- Miura, Y.; Miura, O.; Ihle, J.N.; Aoki, N. Activation of the Mitogen-Activated Protein Kinase Pathway by the Erythropoietin Receptor. J. Biol. Chem. 1994, 269, 29962–29969. [Google Scholar] [CrossRef]
- Arai, A.; Kanda, E.; Nosaka, Y.; Miyasaka, N.; Miura, O. CrkL Is Recruited through Its SH2 Domain to the Erythropoietin Receptor and Plays a Role in Lyn-Mediated Receptor Signaling. J. Biol. Chem. 2001, 276, 33282–33290. [Google Scholar] [CrossRef] [Green Version]
- Mason, J.M.; Beattie, B.K.; Liu, Q.; Dumont, D.J.; Barber, D.L. The SH2 Inositol 5-Phosphatase Ship1 Is Recruited in an SH2-Dependent Manner to the Erythropoietin Receptor. J. Biol. Chem. 2000, 275, 4398–4406. [Google Scholar] [CrossRef] [Green Version]
- Tauchi, T.; Damen, J.E.; Toyama, K.; Feng, G.S.; Broxmeyer, H.E.; Krystal, G. Tyrosine 425 within the Activated Erythropoietin Receptor Binds Syp, Reduces the Erythropoietin Required for Syp Tyrosine Phosphorylation, and Promotes Mitogenesis. Blood 1996, 87, 4495–4501. [Google Scholar] [CrossRef]
- Lee, J.T.; McCubrey, J.A. The Raf/Mek/Erk Signal Transduction Cascade as a Target for Chemotherapeutic Intervention in Leukemia. Leukemia 2002, 16, 486–507. [Google Scholar] [CrossRef] [PubMed]
- Cargnello, M.; Roux, P.P. Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef] [Green Version]
- Pearson, G.; Robinson, F.; Gibson, T.B.; Xu, B.E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-Activated Protein (MAP) Kinase Pathways: Regulation and Physiological Functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar] [CrossRef]
- Wu, P.K.; Becker, A.; Park, J.I. Growth Inhibitory Signaling of the Raf/Mek/Erk Pathway. Int. J. Mol. Sci. 2020, 21, 5436. [Google Scholar] [CrossRef]
- Maik-Rachline, G.; Hacohen-Lev-Ran, A.; Seger, R. Nuclear Erk: Mechanism of Translocation, Substrates, and Role in Cancer. Int. J. Mol. Sci. 2019, 20, 1194. [Google Scholar] [CrossRef] [Green Version]
- Elorza, A.; Hyde, B.; Mikkola, H.K.; Collins, S.; Shirihai, O.S. UCP2 Modulates Cell Proliferation through the MAPK/ERK Pathway during Erythropoiesis and Has No Effect on Heme Biosynthesis. J. Biol. Chem. 2008, 283, 30461–30470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Sytkowski, A.J. Erythropoietin Regulation of Raf-1 and MEK: Evidence for a Ras-Independent Mechanism. Blood 2004, 104, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Kumkhaek, C.; Aerbajinai, W.; Liu, W.; Zhu, J.; Uchida, N.; Kurlander, R.; Hsieh, M.M.; Tisdale, J.F.; Rodgers, G.P. MASL1 Induces Erythroid Differentiation in Human Erythropoietin-Dependent CD34+ Cells through the Raf/MEK/ERK Pathway. Blood 2013, 121, 3216–3227. [Google Scholar] [CrossRef] [Green Version]
- Kondoh, K.; Nishida, E. Regulation of MAP Kinases by MAP Kinase Phosphatases. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 1227–1237. [Google Scholar] [CrossRef] [Green Version]
- Mitin, N.; Rossman, K.L.; Der, C.J. Signaling Interplay in Ras Superfamily Function. Curr. Biol. 2005, 15, 563–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeung, K.; Seitz, T.; Li, S.; Janosch, P.; McFerran, B.; Kaiser, C.; Fee, F.; Katsanakis, K.D.; Rose, D.W.; Mischak, H.; et al. Suppression of Raf-1 Kinase Activity and MAP Kinase Signalling by RKIP. Nature 1999, 401, 173–177. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, A. Regulation of Cytokine Signaling by the SOCS and Spred Family Proteins. Keio J. Med. 2009, 58, 73–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Dev, A.; Verma, R.; Pradeep, A.; Sathyanarayana, P.; Green, J.M.; Narayanan, A.; Wojchowski, D.M. Defining an EPOR- Regulated Transcriptome for Primary Progenitors, Including Tnfr-Sf13c as a Novel Mediator of EPO- Dependent Erythroblast Formation. PLoS ONE 2012, 7, e38530. [Google Scholar] [CrossRef]
- Van Wijk, R.; Van Solinge, W.W. The Energy-Less Red Blood Cell Is Lost: Erythrocyte Enzyme Abnormalities of Glycolysis. Blood 2005, 106, 4034–4042. [Google Scholar] [CrossRef] [PubMed]
- Yudin, J.; Verhovsek, M. How we diagnose and manage altered oxygen affinity hemoglobin variants. Am. J. Hematol. 2019, 94, 597–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thom, C.S.; Dickson, C.F.; Gell, D.A.; Weiss, M.J. Hemoglobin Variants: Biochemical Properties and Clinical Correlates. Cold Spring Harb. Perspect. Med. 2013, 3, a011858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunn, H.F. Erythropoietin. Cold Spring Harb. Perspect. Med. 2013, 3, a011619. [Google Scholar] [CrossRef] [Green Version]
- Tashi, T.; Song, J.; Prchal, J.T. Congenital and Evolutionary Modulations of Hypoxia Sensing and Their Erythroid Phenotype. Curr. Opin. Physiol. 2019, 7, 27–32. [Google Scholar] [CrossRef]
- Vora, S.; Corash, L.; Engel, W.K.; Durham, S.; Seaman, C.; Piomelli, S. The Molecular Mechanism of the Inherited Phosphofructokinase Deficiency Associated with Hemolysis and Myopathy. Blood 1980, 55, 629–635. [Google Scholar] [CrossRef] [Green Version]
- Kristan, A.; Pajič, T.; Maver, A.; Režen, T.; Kunej, T.; Količ, R.; Vuga, A.; Fink, M.; Žula, Š.; Podgornik, H.; et al. Identification of Variants Associated with Rare Hematological Disorder Erythrocytosis Using Targeted Next-Generation Sequencing Analysis. Front. Genet. 2021, 12, 232. [Google Scholar] [CrossRef]
- Katsumura, K.R.; DeVilbiss, A.W.; Pope, N.J.; Johnson, K.D.; Bresnick, E.H. Transcriptional mechanisms underlying hemoglobin synthesis. Cold Spring Harb. Perspect. Med. 2013, 3, a015412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mairbaurl, H.; Humpeler, E. The Influence of Noradrenaline on the Oxygen Affinity of Hemoglobin. Pflugers Arch. Eur. J. Physiol. 1979, 48, 327–386. [Google Scholar]
- Fan, A.X.; Hossain, M.A.; Stees, J.; Gavrilova, E.; Bungert, J. Regulation of erythroid cell differentiation by transcription factors, chromatin structure alterations, and noncoding RNA. In Epigenetic Gene Expression and Regulation; Huang, S., Litt, M.D., Blakey, C., Eds.; Academic Press: Cambridge, MA, USA, 2015; p. 482. [Google Scholar]
- Forget, B.G.; Franklin Bunn, H. Classification of the Disorders of Hemoglobin. Cold Spring Harb. Perspect. Med. 2013, 3, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moradkhani, K.; Préhu, C.; Old, J.; Henderson, S.; Balamitsa, V.; Luo, H.Y.; Poon, M.C.; Chui, D.H.K.; Wajcman, H.; Patrinos, G.P. Mutations in the Paralogous Human α-Globin Genes Yielding Identical Hemoglobin Variants. Ann. Hematol. 2009, 88, 535–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinberg, M.H.; Rodgers, G.P. HbA2: Biology, Clinical Relevance and a Possible Target for Ameliorating Sickle Cell Disease. Br. J. Haematol. 2015, 170, 781–787. [Google Scholar] [CrossRef]
- Nkya, S.; Nkya, S.; Mwita, L.; Mgaya, J.; Kumburu, H.; Van Zwetselaar, M.; Menzel, S.; Mazandu, G.K.; Mazandu, G.K.; Mazandu, G.K.; et al. Identifying Genetic Variants and Pathways Associated with Extreme Levels of Fetal Hemoglobin in Sickle Cell Disease in Tanzania. BMC Med. Genet. 2020, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Thein, S.L.; Menzel, S. Discovering the Genetics Underlying Foetal Haemoglobin Production in Adults. Br. J. Haematol. 2009, 145, 455–467. [Google Scholar] [CrossRef]
- McMullin, M.F. The Classification and Diagnosis of Erythrocytosis. Int. J. Lab. Hematol. 2008, 30, 447–459. [Google Scholar] [CrossRef]
- De Rosa, M.C.; Alinovi, C.C.; Galtieri, A.; Scatena, R.; Giardina, B. The Plasma Membrane of Erythrocytes Plays a Fundamental Role in the Transport of Oxygen, Carbon Dioxide and Nitric Oxide and in the Maintenance of the Reduced State of the Heme Iron. Gene 2007, 398, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, J.D.; Allen, S.L.; Beutler, E.; Kubik, K.; West, C.; Fairbanks, V.F. Erythrocytosis Due to Bisphosphoglycerate Mutase Deficiency with Concurrent Glucose-6-Phosphate Dehydrogenase (G-6-PD) Deficiency. Am. J. Hematol. 2004, 75, 205–208. [Google Scholar] [CrossRef]
- Endeward, V.; Cartron, J.; Ripoche, P.; Gros, G. RhAG Protein of the Rhesus Complex Is a CO2 Channel in the Human Red Cell Membrane. FASEB J. 2008, 22, 64–73. [Google Scholar] [CrossRef]
- Sun, K.; Zhang, Y.; D’Alessandro, A.; Nemkov, T.; Song, A.; Wu, H.; Liu, H.; Adebiyi, M.; Huang, A.; Wen, Y.E.; et al. Sphingosine-1-Phosphate Promotes Erythrocyte Glycolysis and Oxygen Release for Adaptation to High-Altitude Hypoxia. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Reithmeier, R.A.F.; Casey, J.R.; Kalli, A.C.; Sansom, M.S.P.; Alguel, Y.; Iwata, S. Band 3, the Human Red Cell Chloride/Bicarbonate Anion Exchanger (AE1, SLC4A1), in a Structural Context. Biochim. Biophys. Acta Biomembr. 2016, 1858, 1507–1532. [Google Scholar] [CrossRef]
- Cho, J.; King, J.S.; Qian, X.; Harwood, A.J.; Shears, S.B. Dephosphorylation of 2,3-Bisphosphoglycerate by MIPP Expands the Regulatory Capacity of the Rapoport-Luebering Glycolytic Shunt. Proc. Natl. Acad. Sci. USA 2008, 105, 5998–6003. [Google Scholar] [CrossRef] [Green Version]
- Rieger, M.; Schroeder, T. Hematopoiesis. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Parisi, S.; Finelli, C.; Fazio, A.; De Stefano, A.; Mongiorgi, S.; Ratti, S.; Cappellini, A.; Billi, A.M.; Cocco, L.; Follo, M.Y.; et al. Clinical and Molecular Insights in Erythropoiesis Regulation of Signal Transduction Pathways in Myelodysplastic Syndromes and β-Thalassemia. Int. J. Mol. Sci. 2021, 22, 827. [Google Scholar] [CrossRef]
- Lang, E.; Lang, F. Triggers, Inhibitors, Mechanisms, and Significance of Eryptosis: The Suicidal Erythrocyte Death. Biomed. Res. Int. 2015, 2015, 513518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosso, R.; Fader, C.; Colombo, M. Autophagy: A Necessary Event during Erythropoiesis. Blood Rev. 2017, 31, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Nairz, M.; Weiss, G. Iron in Health and Disease. Mol. Asp. Med. 2020, 75, 100906. [Google Scholar] [CrossRef]
- Ginzburg, Y.Z.; Feola, M.; Zimran, E.; Varkonyi, J.; Ganz, T.; Hoffman, R. Dysregulated Iron Metabolism in Polycythemia Vera: Etiology and Consequences. Leukemia 2018, 32, 2105–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganz, T.; Nemeth, E. Hepcidin and Iron Homeostasis. Biochim. Biophys. Acta 2014, 1823, 1434–1443. [Google Scholar] [CrossRef] [Green Version]
- Anderson, E.R.; Xue, X.; Shah, Y.M. Intestinal Hypoxia-Inducible Factor-2α (HIF-2α) Is Critical for Efficient Erythropoiesis. J. Biol. Chem. 2011, 286, 19533–19540. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, A.J.; Das, N.K.; Ramakrishnan, S.K.; Jain, C.; Jurkovic, M.T.; Wu, J.; Nemeth, E.; Lakhal-Littleton, S.; Colacino, J.A.; Shah, Y.M. Hepatic Hepcidin/Intestinal HIF-2α Axis Maintains Iron Absorption during Iron Deficiency and Overload. J. Clin. Investig. 2019, 129, 336–348. [Google Scholar] [CrossRef]
- Forejtnikovà, H.; Vieillevoye, M.; Zermati, Y.; Lambert, M.; Pellegrino, R.M.; Guihard, S.; Gaudry, M.; Camaschella, C.; Lacombe, C.; Roetto, A.; et al. Transferrin Receptor 2 Is a Component of the Erythropoietin Receptor Complex and Is Required for Efficient Erythropoiesis. Blood 2010, 116, 5357–5367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grootendorst, S.; de Wilde, J.; van Dooijeweert, B.; van Vuren, A.; van Solinge, W.; Schutgens, R.; van Wijk, R.; Bartels, M. The Interplay between Drivers of Erythropoiesis and Iron Homeostasis in Rare Hereditary Anemias: Tipping the Balance. Int. J. Mol. Sci. 2021, 22, 2204. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Qu, A.; Anderson, E.R.; Matsubara, T.; Martin, A.; Gonzalez, F.J.; Shah, Y.M. Hypoxia-Inducible Factor-2α Mediates the Adaptive Increase of Intestinal Ferroportin during Iron Deficiency in Mice. Gastroenterology 2011, 140, 2044–2055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, M.C.; Zhang, D.L.; Ollivierre, H.; Eckhaus, M.A.; Rouault, T.A. Translational Repression of HIF2α Expression in Mice with Chuvash Polycythemia Reverses Polycythemia. J. Clin. Investig. 2018, 128, 1317–1325. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, N.; Pantopoulos, K. IRP1 Regulates Erythropoiesis and Systemic Iron Homeostasis by Controlling HIF2α MRNA Translation. Blood 2013, 122, 1658–1669. [Google Scholar] [CrossRef] [Green Version]
- Burlet, B.; Bourgeois, V.; Buriller, C.; Aral, B.; Airaud, F.; Gardie, B.; Girodon, F. High HFE mutation incidence in idiopathic erythrocytosis. Br. J. Haematol. 2019, 185, 794–795. [Google Scholar] [CrossRef] [Green Version]
- Grisouard, J.; Li, S.; Kubovcakova, L.; Rao, T.N.; Meyer, S.C.; Lundberg, P.; Hao-Shen, H.; Romanet, V.; Murakami, M.; Radimerski, T.; et al. JAK2 Exon 12 Mutant Mice Display Isolated Erythrocytosis and Changes in Iron Metabolism Favoring Increased Erythropoiesis. Blood 2016, 128, 839–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Twitchell, D.K.; Pastuszak, A.W.; Khera, M. Controversies in Testosterone Therapy. Sex. Med. Rev. 2021, 9, 149–159. [Google Scholar] [CrossRef]
- Bunn, H.F.; Gu, J.; Huang, L.E.; Park, J.W.; Zhu, H. Erythropoietin: A model system for studying oxygen-dependent gene regulation. J. Exp. Biol. 1998, 201 Pt 8, 1197–1201. [Google Scholar] [CrossRef]
- Tuschl, K.; Clayton, P.T.; Gospe, S.M.; Gulab, S.; Ibrahim, S.; Singhi, P.; Aulakh, R.; Ribeiro, R.T.; Barsottini, O.G.; Zaki, M.S.; et al. Syndrome of hepatic cirrhosis, dystonia, polycythemia, and hypermanganesemia caused by mutations in SLC30A10, a manganese transporter in man. Am. J. Hum. Genet. 2012, 90, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Knight, T.; Urooj Zaidi, A.; Wu, S.; Gadgeel, M.; Buck, S.; Ravindranath, Y. Mild erythrocytosis as a presenting manifestation of PIEZO1 associated erythrocyte volume disorders. Pediatr. Hematol. Oncol. 2019, 36, 317–326. [Google Scholar] [CrossRef]
- Filser, M.; Giansily-Blaizot, M.; Grenier, M. Increased incidence of germline PIEZO1 mutations in individuals with idiopathic erythrocytosis. Blood 2021, 137, 1828–1832. [Google Scholar] [CrossRef]
- Kiger, L.O.; Guitton, C.; Bendélac, L.; Ghazal, K.; Proulle, V.; Galacteros, F.; Junot, C.; Fenaille, F.; Roméo, P.H.; Garçon, L.; et al. Piezo1-xerocytosis red cell metabolome shows impaired glycolysis and increased hemoglobin oxygen affinity. Laurent. Blood Adv. 2021, 5, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Dubin, A.E.; Zhang, Y.; Mousavi, S.A.R.; Wang, Y.; Coombs, A.M.; Loud, M.; Andolfo, I.; Patapoutian, A. A role of PIEZO1 in iron metabolism in mice and humans. Cell 2021, 184, 969–982. [Google Scholar] [CrossRef]
- Jankovsky, N.; Caulier, A.; Demagny, J.; Guitton, C.; Djordjevic, S.; Lebon, D.; Ouled-Haddou, H.; Picard, V.; Garçon, L. Recent advances in the pathophysiology of PIEZO1-relatedhereditary xerocytosis. Am. J. Hematol. 2021, 96, 1017–1026. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomc, J.; Debeljak, N. Molecular Pathways Involved in the Development of Congenital Erythrocytosis. Genes 2021, 12, 1150. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12081150
Tomc J, Debeljak N. Molecular Pathways Involved in the Development of Congenital Erythrocytosis. Genes. 2021; 12(8):1150. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12081150
Chicago/Turabian StyleTomc, Jana, and Nataša Debeljak. 2021. "Molecular Pathways Involved in the Development of Congenital Erythrocytosis" Genes 12, no. 8: 1150. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12081150