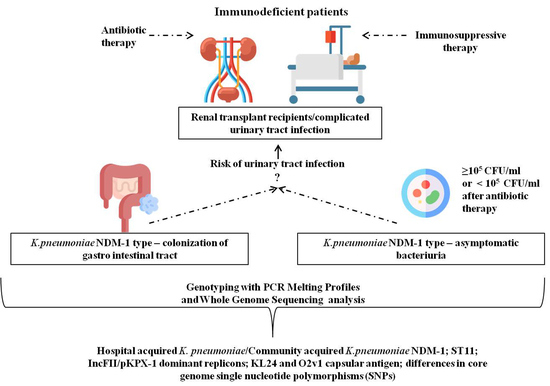
Genetic Background and Antibiotic Resistance Profiles of K. pneumoniae NDM-1 Strains Isolated from UTI, ABU, and the GI Tract, from One Hospital in Poland, in Relation to Strains Nationally and Worldwide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and K. pneumoniae Collection
2.2. Antibiotic Susceptibility Testing
2.3. Ethical Statements
2.4. DNA Isolation
2.5. Clonal Genetic Relatedness Analysis by PCR MP Genotyping
2.6. Whole Genome Sequencing (WGS) and Genome Assembling
2.7. Phylogenetic Analysis of K. pneumoniae Genomes
2.8. Single Nucleotide Polymorphism (SNP) Identification
2.9. Virulence Factors and Resistance Gene Analysis
2.10. K/O Serotypes and Mobile Genetic Element Analysis
2.11. Scoring Contigs as Plasmid or Chromosomal from Draft Genomes
2.12. Data Availability
3. Results
3.1. Patients and Samples from the Local Hospital
3.2. K. pneumoniae Antibiotic Susceptibility Profile
3.3. Clonality of Strains from a Local Outbreak
3.4. Whole Genome Sequence Analysis
3.4.1. Phylogenetic Analysis
3.4.2. SNP Diversity of Core Genomes
3.4.3. Resistomes
3.4.4. Plasmid Analysis
3.4.5. Mobile Genetic Elements Analysis
3.4.6. Serotypes
3.4.7. Virulomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae Population Genomics and Antimicrobial-Resistant Clones. Trends Microbiol. 2016, 24, 944–956. [Google Scholar] [CrossRef] [Green Version]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albiger, B.; Glasner, C.; Struelens, M.J.; Grundmann, H.; Monnet, D.L. European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) working group. Carbapenemase-producing Enterobacteriaceae in Europe: Assessment by national experts from 38 countries, May 2015. Eurosurveillance 2015, 20, 30062. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-β-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Deshpande, P.; Rodrigues, C.; Shetty, A.; Kapadia, F.; Hedge, A.; Soman, R. New Delhi metallo-β lactamase (NDM-1) in Enterobacteriaceae: Treatment options with carbapenems compromised. J. Assoc. Physicians India 2010, 58, 147–149. [Google Scholar] [PubMed]
- Juhas, M.; van der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Datta, S.; Roy, S.; Ramanan, L.; Saha, A.; Viswanathan, R.; Som, T. Carbapenem Resistance in Acinetobacter baumannii and Other Acinetobacter spp. causing neonatal sepsis: Focus on NDM-1 and Its Linkage to ISAba125. Front. Microbiol. 2016, 7, 1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, L.J.; Bahr, G.; Nakashige, T.G.; Nolan, E.M.; Bonomo, R.A.; Vila, A.J. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat. Chem. Biol. 2016, 12, 516–522. [Google Scholar] [CrossRef] [Green Version]
- Bahr, G.; Vitor-Horen, L.; Bethel, C.R.; Bonomo, R.A.; González, L.J.; Vila, A.J. Clinical Evolution of New Delhi Metallo-β-Lactamase (NDM) Optimizes resistance under Zn(II) deprivation. Antimicrob. Agents Chemother. 2017, 62, e01849–e01917. [Google Scholar] [CrossRef] [Green Version]
- Fuursted, K.; Schøler, L.; Hansen, F.; Dam, K.; Bojer, M.S.; Hammerum, A.M.; Dagnæs-Hansen, F.; Olsen, A.; Jasemian, Y.; Struve, C. Virulence of a Klebsiella pneumoniae strain carrying the New Delhi metallo-β-lactamase-1 (NDM-1). Microbes Infect. 2012, 14, 155–158. [Google Scholar] [CrossRef]
- Martin, R.M.; Cao, J.; Wu, W.; Zhao, L.; Manthei, D.M.; Pirani, A.; Snitkin, E.; Malani, P.N.; Rao, K.; Bachman, M.A. Identification of pathogenicity-associated loci in Klebsiella pneumoniae from hospitalized patients. Msystems 2018, 3, e00015–e00018. [Google Scholar] [CrossRef] [Green Version]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.J.; Lin, T.L.; Chen, C.T.; Chen, Y.Y.; Hsieh, P.F.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 2015, 5, 15573. [Google Scholar] [CrossRef] [Green Version]
- Walker, K.A.; Miner, T.A.; Palacios, M.; Trzilova, D.; Frederick, D.R.; Broberg, C.A.; Sepúlveda, V.E.; Quinn, J.D.; Miller, V.L. Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. MBio 2019, 26, e00089–e00119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wacharotayankun, R.; Arakawa, Y.; Ohta, M.; Tanaka, K.; Akashi, T.; Mori, M.; Kato, N. Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect. Immun. 1993, 61, 3164–3174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NCBI Pathogen Detection Reference Gene Catalog. BlaNDM. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pathogens/isolates#/refgene/blaNDM (accessed on 20 June 2020).
- Baraniak, A.; Izdebski, R.; Fiett, J.; Gawryszewska, I.; Bojarska, K.; Herda, M.; Literacka, E.; Żabicka, D.; Tomczak, H.; Pewińska, N.; et al. NDM-producing Enterobacteriaceae in Poland, 2012-14: Inter-regional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. J. Antimicrob. Chemother. 2016, 71, 85–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef] [PubMed]
- Moubareck, C.A.; Mouftah, S.F.; Pál, T.; Ghazawi, A.; Halat, D.H.; Nabi, A.; AlSharhan, M.A.; AlDeesi, Z.O.; Peters, C.C.; Celiloglu, H.; et al. Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int. J. Antimicrob. Agents 2018, 52, 90–95. [Google Scholar] [CrossRef]
- Chen, L.; Mathema, B.; Chavda, K.D.; DeLeo, F.R.; Bonomo, R.A.; Kreiswirth, B.N. Carbapenemase-producing Klebsiella pneumoniae: Molecular and genetic decoding. Trends Microbiol. 2014, 22, 686–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giske, C.G.; Fröding, I.; Hasan, C.M.; Turlej-Rogacka, A.; Toleman, M.; Livermore, D.; Woodford, N.; Walsh, T.R. Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob. Agents Chemother. 2012, 56, 2735–2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baraniak, A.; Machulska, M.; Żabicka, D.; Literacka, E.; Izdebski, R.; Urbanowicz, P.; Bojarska, K.; Herda, M.; Kozińska, A.; Hryniewicz, W.; et al. Towards endemicity: Large-scale expansion of the NDM-1-producing Klebsiella pneumoniae ST11 lineage in Poland, 2015-16. J. Antimicrob. Chemother. 2019, 74, 3199–3204. [Google Scholar] [CrossRef] [PubMed]
- Żabicka, D.; Bojarska, K.; Herda, M.; Literacka, E.; Kozińska, A.; Hryniewicz, W.; Skoczyńska, A.; Baraniak, A.; Machulska, M.; Urbanowicz, P.; et al. Pałeczki Jelitowe Enterobacteriaceae Wytwarzające Karbapenemazy (CPE) w Polsce-Sytuacja w 2016. Krajowy Ośrodek Referencyjny ds. Lekowrażliwości Drobnoustrojów. Available online: http://www.korld.edu.pl/pdf/CPEraport2016.pdf (accessed on 25 November 2017).
- Żabicka, D.; Literacka, E.; Gniadkowski, M.; Hryniewicz, W. Raport Krajowego Ośrod-ka Referencyjnego ds. Lekowrażliwości Drobnoustrojów. Występowanie Entero- bacteriaceae (głównie Klebsiella pneumoniae), wytwarzających karbapenemazę New Delhi (NDM) na terenie Polski w okresie I–III kwartał 2017 roku. KORLD. Available online: http://www.korld.edu.pl/pdf/Raport_NDM_18-12-2017 (accessed on 20 December 2017).
- Karuthu, S.; Blumberg, E.A. Common infections in kidney transplant recipients. Clin. J. Am. Soc. Nephrol. 2012, 7, 2058–2070. [Google Scholar] [CrossRef] [Green Version]
- Ko, K.S.; Cho, D.O.; Ahn, J.H.; Lee, T.W.; Ihm, C.G.; Chang, S.G.; Chai, S.E.; Park, H.C.; Hong, S.H.; Joo, H.Z.; et al. Infections after renal transplantation. Transplant. Proc. 1994, 26, 2072–2074. [Google Scholar] [PubMed]
- Säemann, M.; Hörl, W.H. Urinary tract infection in renal transplant recipients. Eur. J. Clin. Invest 2008, 38 (Suppl. S2), 58–65. [Google Scholar] [CrossRef]
- Vidal, E.; Torre-Cisneros, J.; Blanes, M.; Montejo, M.; Cervera, C.; Aguado, J.M.; Len, O.; Carratalá, J.; Cordero, E.; Bou, G.; et al. Bacterial urinary tract infection after solid organ transplantation in the RESITRA cohort. Transpl. Infect. Dis. 2012, 14, 595–603. [Google Scholar] [CrossRef]
- Izdebski, R.; Sitkiewicz, M.; Urbanowicz, P.; Krawczyk, M.; Brisse, S.; Gniadkowski, M. Genomic background of the Klebsiella pneumoniae NDM-1 outbreak in Poland, 2012–18. J. Antimicrob. Chemother. 2020, 75, 3156–3162. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Nordmann, P.; Poirel, L.; Carrër, A.; Toleman, M.A.; Walsh, T.R. How to detect NDM-1 producers. J. Clin. Microbiol. 2011, 49, 718–721. [Google Scholar] [CrossRef] [Green Version]
- Stojowska, K.; Kałuzewski, S.; Krawczyk, B. Usefulness of PCR melting profile method for genotyping analysis of Klebsiella oxytoca isolates from patients of a single hospital unit. Pol. J. Microbiol. 2009, 58, 247–253. [Google Scholar]
- Gołębiewska, J.E.; Krawczyk, B.; Wysocka, M.; Ewiak, A.; Komarnicka, J.; Bronk, M.; Rutkowski, B.; Dębska-Ślizień, A. Host and pathogen factors in Klebsiella pneumoniae upper urinary tract infections in renal transplant patients. J. Med. Microbiol. 2019, 68, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling genomes and mini-metagenomes from highly chimeric reads. In Research in Computational Molecular Biology. RECOMB 2013. Lecture Notes in Computer Science; Deng, M., Jiang, R., Sun, F., Zhang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 7821, pp. 158–170. [Google Scholar]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T. Ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Tonkin-Hill, G.; Lees, J.A.; Bentley, S.D.; Frost, S.D.W.; Corander, J. RhierBAPS: An R implementation of the population clustering algorithm hierBAPS. Wellcome Open Res. 2018, 3, 93. [Google Scholar] [CrossRef] [Green Version]
- MLST. Available online: https://github.com/tseemann/mlst (accessed on 25 June 2020).
- Snippy, Rapid Haploid Variant Calling and Core Genome Alignment. Available online: https://github.com/tseemann/snippy (accessed on 25 June 2020).
- SnpEff. Genomic Variant Annotations and Functional Effect Prediction Toolbox. Available online: http://snpeff.sourceforge.net (accessed on 25 June 2020).
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Wick, R.R.; Heinz, E.; Holt, K.E.; Wyres, K.L. Kaptive Web: User-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. BioRxiv 2018, 56, e00197–e00218. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2018. Available online: http://www.R-project.org/ (accessed on 19 July 2021).
- Farzand, R.; Rajakumar, K.; Zamudio, R.; Oggioni, M.R.; Barer, M.R.; O’Hare, H.M. ICEKp2: Description of an integrative and conjugative element in Klebsiella pneumoniae, co-occurring and interacting with ICEKp1. Sci. Rep. 2019, 9, 13892. [Google Scholar] [CrossRef] [Green Version]
- RFPlasmid. Available online: https://github.com/aldertzomer/RFPlasmid (accessed on 25 June 2020).
- Ko, K.S. The contribution of capsule polysaccharide genes to virulence of Klebsiella pneumoniae. Virulence 2017, 8, 485–486. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.Y.; Chen, Y.S.; Wu, C.Y.; Chang, H.Y.; Lai, Y.C.; Peng, H.L. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 2010, 192, 3144–3158. [Google Scholar] [CrossRef] [Green Version]
- El Fertas-Aissani, R.; Messai, Y.; Alouache, S.; Bakour, R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol. Biol. 2013, 61, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Gołębiewska, J.; Dębska-Ślizień, A.; Komarnicka, J.; Samet, A.; Rutkowski, B. Urinary tract infections in renal transplant recipients. Transplant. Proc. 2011, 43, 2985–2990. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef] [Green Version]
- Taglietti, F.; Di Bella, S.; Galati, V.; Topino, S.; Iappelli, M.; Petrosillo, N. Carbapenemase-producing Klebsiella pneumoniae-related mortality among solid organ-transplanted patients: Do we know enough? Transpl. Infect. Dis. 2013, 15, E164–E165. [Google Scholar] [CrossRef]
- Wilkowski, P.; Ciszek, M.; Dobrzaniecka, K.; Sańko-Resmer, J.; Łabuś, A.; Grygiel, K.; Grochowiecki, T.; Młynarczyk, G.; Pączek, L. Successful treatment of urinary tract infection in kidney transplant recipients caused by multiresistant Klebsiella pneumoniae producing New Delhi metallo-β-lactamase (NDM-1) with strains genotyping. Transplant. Proc. 2016, 48, 1576–1579. [Google Scholar] [CrossRef]
- Linares, L.; Cervera, C.; Hoyo, I.; Sanclemente, G.; Marco, F.; Cofán, F.; Ricart, M.J.; Navasa, M.; Moreno, A. Klebsiella pneumoniae infection in solid organ transplant recipients: Epidemiology and antibiotic resistance. Transplant. Proc. 2010, 42, 2941–2943. [Google Scholar] [CrossRef]
- Zdziarski, J.; Brzuszkiewicz, E.; Wullt, B.; Liesegang, H.; Biran, D.; Voigt, B.; Grönberg-Hernandez, J.; Ragnarsdottir, B.; Hecker, M.; Ron, E.Z.; et al. Host imprints on bacterial genomes--rapid, divergent evolution in individual patients. PLoS Pathog. 2010, 6, e1001078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stork, C.; Kovács, B.; Rózsai, B.; Putze, J.; Kiel, M.; Dorn, Á.; Kovács, J.; Melegh, S.; Leimbach, A.; Kovács, T.; et al. Characterization of asymptomatic bacteriuria Escherichia coli isolates in search of alternative strains for efficient bacterial interference against uropathogens. Front. Microbiol. 2018, 9, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM metallo-β-lactamases and their bacterial producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115–e00118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Liu, Y.; Wang, R.; Wang, Q.; Jin, L.; Wang, H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine 2020, 51, 102599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, L.; Merciecca, T.; Forestier, C.; Balestrino, D.; Miquel, S. From Klebsiella pneumoniae Colonization to Dissemination: An Overview of Studies Implementing Murine Models. Microorganisms 2021, 9, 1282. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Toleman, M.A.; Hansen, F.; Kristensen, B.; Lester, C.H.; Walsh, T.R.; Fuursted, K. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect. Dis. 2010, 10, 829–830. [Google Scholar] [CrossRef]
- Bonomo, R.A. New Delhi metallo-β-lactamase and multidrug resistance: A global SOS? Clin. Infect. Dis. 2011, 52, 485–487. [Google Scholar] [CrossRef] [Green Version]
- Pitout, J.D.; Nordmann, P.; Poirel, L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 2015, 59, 5873–5884. [Google Scholar] [CrossRef] [Green Version]
- Ruppé, E.; Armand-Lefèvre, L.; Estellat, C.; El-Mniai, A.; Boussadia, Y.; Consigny, P.H.; Girard, P.M.; Vittecoq, D.; Bouchaud, O.; Pialoux, G.; et al. Acquisition of carbapenemase-producing Enterobacteriaceae by healthy travellers to India, France, February 2012 to March 2013. Eurosurveillance 2014, 19, 20768. [Google Scholar] [CrossRef]
- Fiett, J.; Baraniak, A.; Izdebski, R.; Sitkiewicz, I.; Żabicka, D.; Meler, A.; Filczak, K.; Hryniewicz, W.; Gniadkowski, M. The first NDM metallo-β-lactamase-producing Enterobacteriaceae isolate in Poland: Evolution of IncFII-type plasmids carrying the bla(NDM-1) gene. Antimicrob. Agents Chemother. 2014, 58, 1203–1207. [Google Scholar] [CrossRef] [Green Version]
- Izdebski, R.; Bojarska, K.; Baraniak, A.; Literacka, E.; Herda, M.; Żabicka, D.; Guzek, A.; Półgrabia, M.; Hryniewicz, W.; Gniadkowski, M. NDM-1- or OXA-48-producing Enterobacteriaceae colonising Polish tourists following a terrorist attack in Tunis, March 2015. Eurosurveillance 2015, 20, 21150. [Google Scholar] [CrossRef]
- Clegg, S.; Murphy, C.N. Epidemiology and virulence of Klebsiella pneumoniae. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Wysocka, M.; Zamudio, R.; Oggioni, M.R.; Gołębiewska, J.; Dudziak, A.; Krawczyk, B. The new Klebsiella pneumoniae ST152 variants with hypermucoviscous phenotype isolated from renal transplant recipients with asymptomatic bacteriuria-genetic characteristics by WGS. Genes 2020, 11, 1189. [Google Scholar] [CrossRef]
- Schroll, C.; Barken, K.B.; Krogfelt, K.A.; Struve, C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010, 10, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcántar-Curiel, M.D.; Blackburn, D.; Saldaña, Z.; Gayosso-Vázquez, C.; Iovine, N.; De la Cruz, M.A.; Girón, J.A. Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence 2013, 4, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Phillips, P.L.; Schultz, G.S. Molecular Mechanisms of Biofilm Infection: Biofilm Virulence Factors. Adv. Wound Care 2012, 1, 109–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawlor, M.S.; O’connor, C.; Miller, V.L. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 2007, 75, 1463–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachman, M.A.; Oyler, J.E.; Burns, S.H.; Caza, M.; Lépine, F.; Dozois, C.M.; Weiser, J.N. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 2011, 79, 3309–3316. [Google Scholar] [CrossRef] [Green Version]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, V.; Klemm, P. Global gene expression profiling of asymptomatic bacteriuria Escherichia coli during biofilm growth in human urine. Infect. Immun. 2007, 75, 966–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, C.; Menezes, J.; Belas, A.; Aboim, C.; Cavaco-Silva, P.; Trigueiro, G.; Gama, L.T.; Pomba, C. Klebsiella pneumoniae causing urinary tract infections in companion animals and humans: Population structure, antimicrobial resistance and virulence genes. J. Antimicrob. Chemother. 2019, 74, 594–602. [Google Scholar] [CrossRef]
- Aguirre-Quiñonero, A.; Martínez-Martínez, L. Non-molecular detection of carbapenemases in Enterobacteriaceae clinical isolates. J. Infect. Chemother. 2017, 23, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Dou, Y.; Zhou, K.; Chen, Y.; Han, L.; Guo, X.; Sun, J. Coexistence of blaOXA-48 and truncated blaNDM-1 on different plasmids in a Klebsiella pneumoniae isolate in China. Front. Microbiol. 2017, 8, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidjabat, H.E.; Townell, N.; Nimmo, G.R.; George, N.M.; Robson, J.; Vohra, R.; Davis, L.; Heney, C.; Paterson, D.L. Dominance of IMP-4-producing Enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob. Agents Chemother. 2015, 59, 4059–4066. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Dortet, L.; Bernabeu, S.; Nordmann, P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 2011, 55, 5403–5407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolain, J.M.; Parola, P.; Cornaglia, G. New Delhi metallo-β-lactamase (NDM-1): Towards a new pandemia? Clin. Microbiol. Infect. 2010, 16, 1699–1701. [Google Scholar] [CrossRef] [Green Version]


| Patients | Isolates | Collection Number | Samples | #Date of Isolation | Result | Renal Transplantation (RTx) |
|---|---|---|---|---|---|---|
| 1 | 1 | ND32 | urine | 2016 | UTI | no |
| 2 | 2 | ND24 | urine | 2018 | ABU | yes |
| 3 | 3 | ND33 | anal swab | 2018 | Colonization/carriage | no |
| 4 | 4 | ND34 | anal swab | 2018 | Colonization/carriage | yes |
| 5 | ND88 | urine | 2018 | ABU | yes | |
| 5 | 6 | ND96 | urine | 2018 | UTI | no |
| 6 | 7 | ND111 | urine | 2018 | ABU | no |
| 7 | 8 * | ND27 | urine | 2018 | UTI | no |
| Antibiotic | Abbreviation | Name of Isolate K. pneumoniae New Delhi | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ND32 | ND24 | ND33 | ND34 | ND88 | ND96 | ND111 | ND27 | ||
| Ampicillin | AMP | R | R | R | R | R | R | R | R |
| Amoxicillin/Clavulanic acid | AMC | R | R | R | R | R | R | R | R |
| Piperacillin/Tazobactam | TZP | R | R | R | R | R | R | R | R |
| Cephalotin | CEF | R | R | R | R | R | R | R | R |
| Cefuroxime | CXM | R | R | R | R | R | R | R | R |
| Cefotaxime | CTX | R | R | NDt | R | R | R | R | R |
| Ceftazidime | CAZ | R | R | R | R | R | R | R | R |
| Cefepime | FEP | R | R | R | R | R | R | R | R |
| Cefoperazone/Subactam | SUL | R | R | R | R | R | R | R | R |
| Ertapenem | ETP | R | R | R | R | R | R | R | R |
| Imipenem | IPM | I | R | R | I | I | R | I | R |
| Meropenem | MEM | R | R | R | R | R | R | R | R |
| Amikacin | AMK | I | R | S | S | I | S | S | S |
| Gentamicin | GEN | NDt | S | S | NDt | NDt | NDt | NDt | NDt |
| Tobramycin | TOB | NDt | R | S | NDt | NDt | NDt | NDt | NDt |
| Tigecycline | TGC | NDt | I | S | NDt | NDt | NDt | NDt | NDt |
| Ciprofloxacin | CIP | R | R | R | R | R | R | R | R |
| Trimethoprim/Sulfamethoxazole | SXT | S | R | R | R | R | R | R | R |
| Colistin | CST | NDt | S | S | NDt | S | NDt | S | NDt |
| Isolate Name | Plasmid Replicon Profile | Acquired Antimicrobial Resistance Genes a | |||||||
|---|---|---|---|---|---|---|---|---|---|
| β-Lactams | Aminoglycosides b | Fluoroquinolones b | Macrolide, Linosamide, Streptogramin Antibiotics | Phenicols | Sulphonamides | Tetracycline | Trimethoprim | ||
| ND32 | FII (pKPX1) | blaNDM-1, blaCTX-M-15,blaOXA-1 | aac (6′)-Ib | - | - | - | - | - | dfrA14 |
| ND111 | FIB (K), FII (K), FII (pKPX1), HI1A (NDM-CIT), HI1B (pNDM-CIT) | blaNDM-1, blaCTX-M-15, blaOXA-1 | aac (6′)-Ib | - | - | catA1, catB4 | sul1 | - | dfrA1, dfrA14 |
| ND27 | FIB (K), FII (K), FII (pKPX1), HI1A (NDM-CIT), HI1B (pNDM-CIT) | blaNDM-1, blaCTX-M-15, blaOXA-1 | aac (6′)-Ib | - | - | catA1 | sul1 | - | dfrA1, dfrA14 |
| ND96 | FIB (K), FII (K), FII (pKPX1), HI1A (NDM-CIT), HI1B (pNDM-CIT) | blaNDM-1, blaCTX-M-15, blaOXA-1 | aac (6′)-Ib | - | - | catA1 | sul1 | - | dfrA1, dfrA14 |
| ND88 | FIB (K), FII (K), FII (pKPX1), HI1A (NDM-CIT), HI1B (pNDM-CIT), X1 | blaNDM-1, blaCTX-M-15, blaOXA-1, blaTEM-1 | aac (6′)-Ib | - | - | catA1, catB4 | sul1 | - | dfrA1, dfrA14 |
| ND24 | FII (Yp), R | blaNDM-1, blaCTX-M-15 | aac (6′)-Ib | - | - | - | sul2 | - | dfrA14 |
| ND33 | FIB (K), FII (K), FII (pKPX1), HI1A (NDM-CIT), HI1B (pNDM-CIT) | blaNDM-1, blaCTX-M-15 | aadA1 | - | - | catA1 | sul1 | - | dfrA1, dfrA14 |
| ND34 | FIB (K), FII (K), FII (pKPX1), HI1A (NDM-CIT), HI1B (pNDM-CIT), X1 | blaNDM-1, blaCTX-M-15, blaOXA-1, blaTEM-1 | aac (6′)-Ib | - | - | catA1 | sul1 | - | dfrA1, dfrA14 |
| KN1960 | C, FIB (K), FIB (pNDM-Mar), FII (K), HI1B (pNDM-MAR), N | blaNDM-1, blaCTX-M-15, blaOXA-1 | aac (6′)-Ib, aac (3)-IIa, aph (3′′)-Ib, aph (3′)-VI, aph (6)-Id | qnrS1 | mph (A) | catA1 | sul1, sul2 | - | dfrA14 |
| KN20454 | FIA (HI1), FIB (K), FII (K), FII (pKPX1), R | blaNDM-1, blaCTX-M-15, blaOXA-1, blaTEM-1 | aac (6′)-Ib, aac (3)-IIa, aph (3′′)-Ib, aph (6)-Id | - | mph (A) | - | sul2 | tet (A) | dfrA14 |
| KN20534 | FIA (HI1), FIB (K), FII (pKPX1), R | blaNDM-1 | aac (6’)-Ib | - | - | - | - | - | dfrA14 |
| KN20537 | Col440II, FIA (HI1), FIB (K), FII (K) | blaNDM-1, blaOXA-1 | aac (6′)-Ib, aac (3)-IIa | - | mph (A) | - | - | tet (A) | - |
| KN20779 | FII (Yp), M2, R | blaNDM-1, blaCTX-M-3, blaOXA-1, blaTEM-1 | aac (6′)-Ib-cr, aac (3)-IId, aadA2, armA | aac (6′)-Ib-cr, qnrB4 | mph (A), mph (E), msr (E) | catB3 | sul1 | tet (A) | dfrA12 |
| KN21238 | FIB (K) (pCAV1099-114), FIB (pQiI), FII (pKPX1), HI2, HI2A | blaNDM-1, blaCTX-M-9 | aadA2 | - | - | catA1 | sul1 | - | dfrA16 |
| KN21460 | FIA (HI1), FIB (K), FII (K), M1, R | blaNDM-1, blaTEM-1 | aac (6′)-Ib, aph (3′′)-Ib, aph (6)-Id | - | mph (A) | - | sul2 | tet (A) | dfrA14 |
| KN6983 | Col440I, FIB (K), FII (K), FII (pKPX1), L | blaNDM-1, blaCTX-M-15, blaOXA-1,-48 | aac (6′)-Ib-cr, aac (3)-IIa, aph (3′)-Ia, aadA2 | aac (6′)-Ib-cr | mph (A) | - | sul1 | - | dfrA12 |
| KN6988 | Col440l, FIB (pKPHS1), FIB (pNDM-Mar), FIB (pQil), HI1B (pNDM-MAR) | blaNDM-1, blaCTX-M-15, blaOXA-1,-10, blaTEM-1 | aac (6′)-Ib-cr, aac (3)-IIa, aph (3′′)-Ib, aph (3′)-VI, aph (6)-Id, armA | aac (6′)-Ib-cr, qnrS1 | msr (E) | catA1 | sul1 | tet (A) | dfrA1 |
| KN20452 | FIA (HI1), FIB (K), FIB (pQiI), FII (K), FII (pKPX1), R | blaNDM-1, blaCTX-M-15, blaOXA-1, blaTEM-1 | aac (6′)-Ib, aac (6′)-Ib-cr, aac (3)-IIa, aph (3′′)-Ib, aph (6)-Id | aac (6′)-Ib-cr | mph (A) | - | sul2 | tet (A) | dfrA14 |
| KN20470 | FIA (HI1), FIB (K), FII (K), FII (pKPX1), R | blaNDM-1, blaCTX-M-15, blaOXA-1, blaTEM-1 | aac (6′)-Ib, aac (3)-IIa, aph (3′′)-Ib, aph (6)-Id | - | mph (A) | - | sul2 | tet (A) | dfrA14 |
| KN20531 | FIA (HI1), FIB (K), FII (K), FII (pKPX1) | blaNDM-1, blaCTX-M-15, blaOXA-1 | aac (6′)-Ib, aac (3)-IIa | - | - | - | - | tet (A) | dfrA14 |
| KN20607 | Col (BS512), FIA (HI1), FIB (K), FII (K), R | blaNDM-1, blaCTX-M-15, blaOXA-1, blaTEM-1 | aac (6′)-Ib, aac (3)-IIa, aph (3′′)-Ib, aph (6)-Id | - | mph (A) | - | sul2 | tet (A) | dfrA14 |
| KN20611 | FIA (HI1), FIB (K), FII (K), FII (pKPX1), R | blaNDM-1, blaCTX-M-15, blaOXA-1, blaTEM-1 | aac (6′)-Ib, aac (6′)-Ib-cr, aac (3)-IIa, aph (3′’)-Ib, aph (6)-Id | aac (6′)-Ib-cr | mph (A) | - | sul2 | tet (A) | dfrA14 |
| KN20617 | FIA (HI1), FIB (K), FII (K), FII (pKPX1), R | blaNDM-1 | aac (6′)-Ib, aph (3′′)-Ib, aph (6)-Id | - | mph (A) | - | sul2 | tet (A) | dfrA14 |
| KN19957 | FIA (HI1), FIB (K), FII (K), FII (Yp), R | blaNDM-1, blaCTX-M-15 | aac (6′)-Ib, aac (3)-IIa | qnrB9 | mph (A) | - | sul1 | tet (A) | . |
| Sample Name | Serotypes | Virulence Factors | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Capsule (K) Antigen Types | LPS (O) Antigen Types | Type 3 Fimbriae | Type 1 Fimbriae | High-Pathogenicity Island: Yersiniabactin | IroA System: Salmochelin Synthesis | Fep-Ent System: Enterobactin | |||||||
| mrkABCDF | mrkHIJ | fimABCDEFGHIK | fyuA | irp1/2 | ybtAEPQSTUX | iroE | fepABCDG | fes | ybdA | entABCEF | |||
| ND32 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ND111 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ND27 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ND96 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ND88 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ND24 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ND33 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ND34 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN1960 | KL14 | O3b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN20454 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN20534 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN20537 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN20779 | KL105 | O2v2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN21238 | KL15 | O4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN21460 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN6983 | KL15 | O4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN6988 | KL103 | O2v1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 |
| KN20452 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN20470 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN20531 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN20607 | KL24 | O2v1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN20611 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN20617 | KL24 | O2v1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KN19957 | KL24 | O2v1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wysocka, M.; Zamudio, R.; Oggioni, M.R.; Gołębiewska, J.; Bronk, M.; Krawczyk, B. Genetic Background and Antibiotic Resistance Profiles of K. pneumoniae NDM-1 Strains Isolated from UTI, ABU, and the GI Tract, from One Hospital in Poland, in Relation to Strains Nationally and Worldwide. Genes 2021, 12, 1285. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12081285
Wysocka M, Zamudio R, Oggioni MR, Gołębiewska J, Bronk M, Krawczyk B. Genetic Background and Antibiotic Resistance Profiles of K. pneumoniae NDM-1 Strains Isolated from UTI, ABU, and the GI Tract, from One Hospital in Poland, in Relation to Strains Nationally and Worldwide. Genes. 2021; 12(8):1285. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12081285
Chicago/Turabian StyleWysocka, Magdalena, Roxana Zamudio, Marco R. Oggioni, Justyna Gołębiewska, Marek Bronk, and Beata Krawczyk. 2021. "Genetic Background and Antibiotic Resistance Profiles of K. pneumoniae NDM-1 Strains Isolated from UTI, ABU, and the GI Tract, from One Hospital in Poland, in Relation to Strains Nationally and Worldwide" Genes 12, no. 8: 1285. https://0-doi-org.brum.beds.ac.uk/10.3390/genes12081285