RNAi in Arthropods: Insight into the Machinery and Applications for Understanding the Pathogen-Vector Interface
Abstract
:1. Introduction
| Vector | Pathogen | Disease | Gene counts |
|---|---|---|---|
| Culex quinquefasciatus (Southern house mosquito) | West Nile virus Wuchereria bancrofti | West Nile Fever Lymphatic filariasis | Genome (bp): 539,959,374 Known protein-coding genes: 18,858 Gene exons: 75,303 Gene transcripts: 23,049 |
| Anopheles gambiae | Plasmodium spp. | Malaria | Genome (bp): 278,253,050 Known protein-coding genes: 12,670 Gene exons: 56,210 Gene transcripts: 14,974 |
| Aedes aegypti (Yellow fever mosquito) | Dengue virus Chikungunya virus (CHIKV)Yellow fever virus | Dengue fever Chikungunya Yellow fever | Genome (bp): 1,310,090,344 Known protein-coding genes: 15,704 Gene exons: 66,827 Gene transcripts: 18,769 |
| Ixodes scapularis (Deer- or blacklegged tick) | Borrelia burgdorferi Borrelia miyamotoi | Lyme Disease Relapsing fever | Base Pairs: 1,388,472,180 Novel protein-coding genes: 20,457 Gene exons: 93,988 Gene transcripts: 24,925 |
| Pediculus humanus corporis (Body louse) | Infestation of lice Rickettsia prowazekii Bartonella Quintana Borrelia recurrentis | Pediculosis Epidemic typhus Trench fever, endocarditis Louse-borne relapsing fever | Base Pairs: 108,367,968 Known protein-coding genes: 10,773 Gene exons: 69,506 Gene transcripts: 10,994 |
| Rhodnius prolixus (Kissing Bug) | Trypanosoma cruzi | Chagas Disease | Base Pairs: 561,474,548 Pseudogenes: 1,148 Gene exons: 184,075 Gene transcripts: 36,307 |
| Glossina morsitans (Tsetse fly) | Trypanosoma brucei | African trypanosomiasis (sleeping sickness) | Base Pairs: 363,107,930 Gene exons: 64,464 Gene transcripts: 12,362 |
| Trypanosoma b. brucei | Animal trypanosomiasis (Nagana) |
2. Invertebrate RNAi Machinery
| Vector of disease/ Protozoan pariste | Class of small silencing RNAs | RNAi machinery | Reference | |||
|---|---|---|---|---|---|---|
| Dicer (Drosha in miRNA pathways) | RISC complex | Transitive amplification | Systemic protein | |||
| Vector | ||||||
| Tsetse fly (Glossinidae) | Unexplored | ND | ND | ND | ND | [15,16,17] |
| Pathogen(s) transmitted | ||||||
| Trypanosoma brucei | siRNA | TbDCL1 (cytoplasm, RNase IIIa) | TbAGO1, TbRIF5 | Unknown | Unknown | [18,19,20,21,22] |
| TbDLC2 (nucleus, RNase IIIb) | TbRIF4, PIWI-tryp | |||||
| Vector | ||||||
| Triatomine or kissing bugs (Triatome/ Reduviidae) | ||||||
| Rhodnius prolixus | Unexplored | ND | ND | ND | ND | [23] |
| Triatomine brasiliensis | Unexplored | ND | ND | ND | ND | [24] |
| Pathogen(s) transmitted: | ||||||
| Trypanosoma cruzi | piRNA | Absent | PIWI-tryp | Absent | Absent | [25] |
| Vector | ||||||
| Ixodid tick | ||||||
| I. scapularis | miRNA | Drc-1 | Ago-1 (PIWI and PAZ domain) | *Epn-1Cele | Rsd-3 | [26] |
| siRNA | Ago-2 (PAZ) | AP-50, Arf72, Clathrin hc, Rab7, CG3911, Cog3, IdICp | ||||
| Pathogen(s) transmitted: | ||||||
| Babesia, Borrelia, Anaplasma | Unexplored | ND | ND | ND | ND | |
| Vector | ||||||
| Mosquitoes | ||||||
| Aedes aegypti / A .albopticus | miRNA | Drosha, Dicer-1 (Pasha, Loqs) | Ago-1 (x 2) | Lack SID-1, but shows systemic response | [27] | |
| siRNA | Dicer-2, R2D2 | Ago-2 (VIG, TSN, Fmr-1) | ||||
| piRNA | Absent | Ago-3, Ago-4 like (x 4), Ago-5 like (x 3) | ||||
| Culex quinquefasciatus | miRNA | Drosha, Dicer-1 (Pasha, Loqs) | Ago-1 | |||
| siRNA | Dicer-2, R2D2 | Ago-2 (x 2) ( TSN, Fmr-1) | ||||
| piRNA | Absent | Ago-3, Ago-4 like (x 3), Ago-5 like (x 3) | ||||
| Anopheles gambiae | miRNA | Drosha, Dicer-1 (Pasha, Loqs) | Ago-1 | |||
| siRNA | Dicer-2, R2D2 | Ago-2 (Fmr-1) | ||||
| piRNA | Absent | Ago-3, Ago-4 like, Ago-5 like | ||||
| Pathogen(s) transmitted: | ||||||
| Plasmodium | Absent | Absent | Absent | Absent | Absent | [28,29] |
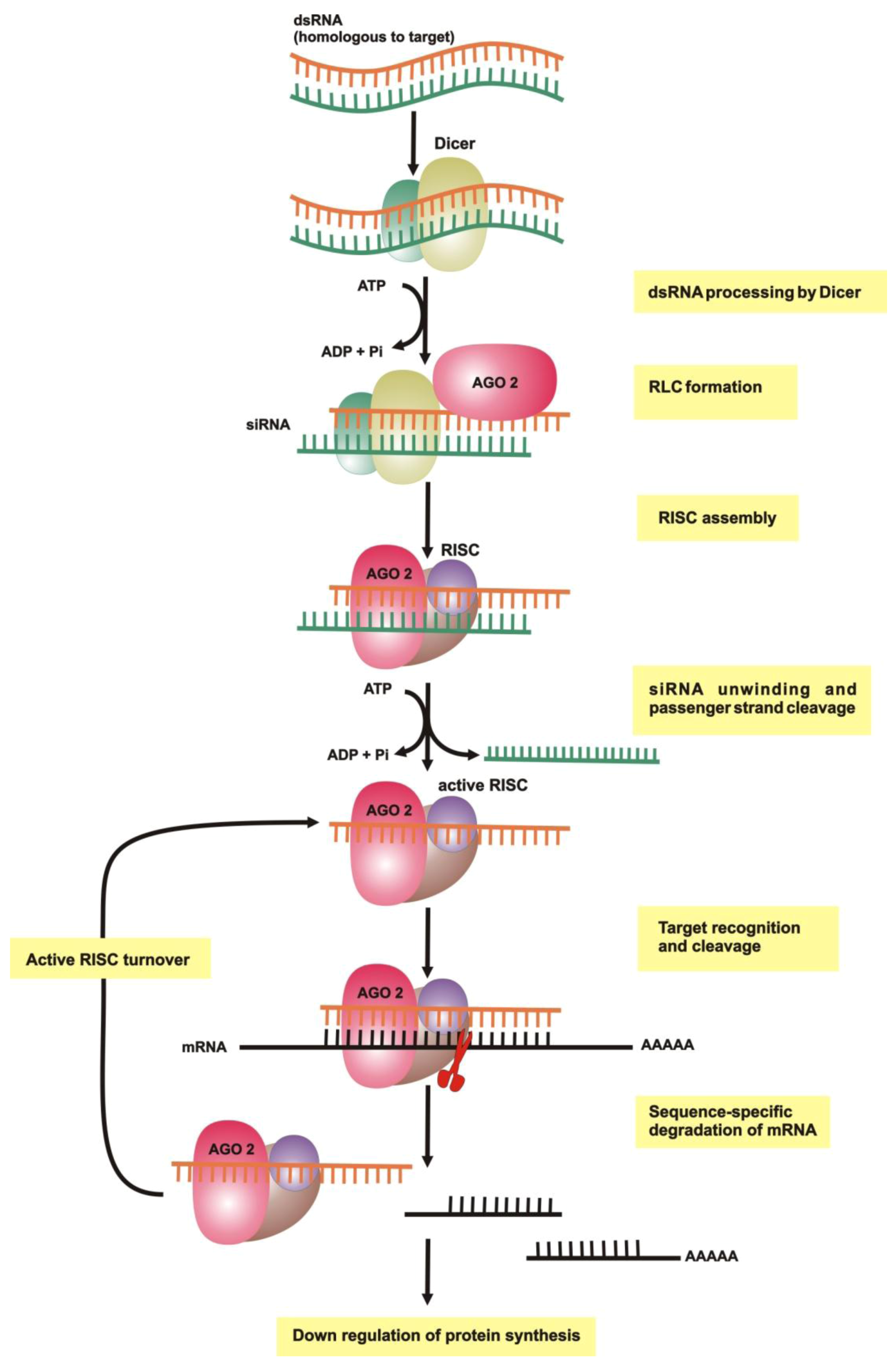
2.1. dsRNA Uptake
2.2. Initiation of RNAi
2.3. RNA-Induced Silencing Complex (RISC) Assembly
2.4. Slicing or Silencing Steps
2.5. Transitive Amplification of the Initial dsRNA Signal
2.6. Systemic RNAi
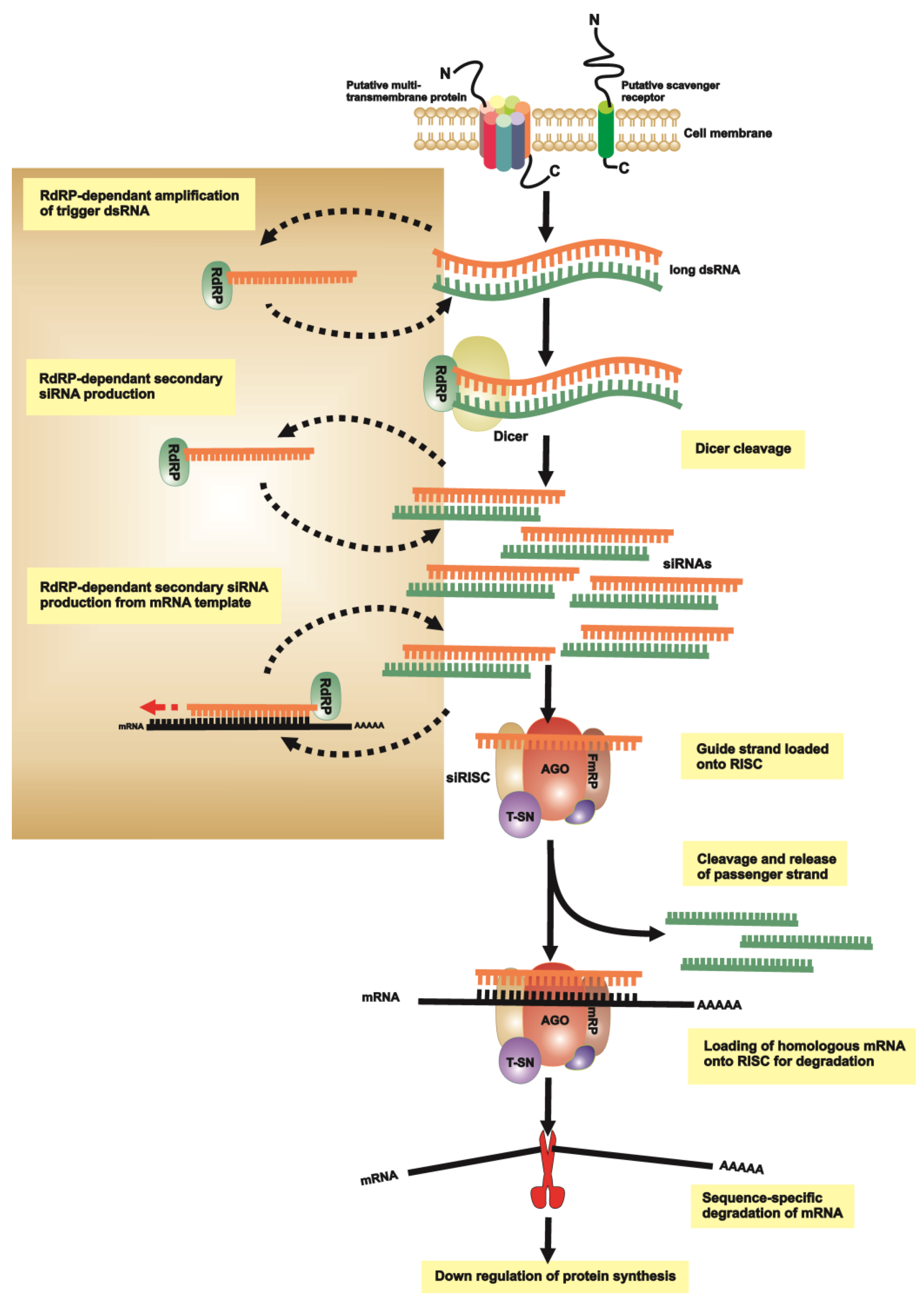
3. RNAi of Lice, Ticks and Tick-Borne Pathogens
3.1. RNAi Pathways and Methodologies
3.2. RNAi to Understand Tick Vector Biology
3.3. RNAi to Understand Tick-Pathogen Interactions
3.4. RNAi in Phthiraptera (Lice)
4. RNAi of Mosquitoes and Their Associated Pathogens
4.1. dsRNA Introduction into Mosquitoes
4.2. RNAi to Understand Mosquito Vector Biology
4.3. RNAi as a Tool for Studying Mosquito Vector-Pathogen Interactions
5. RNAi in Triatominae and Trypanosoma cruzi.
6. RNAi in the Tsetse Fly and Trypanosoma brucei
6.1. RNAi Contributions to Understanding Vector Biology
6.2. RNAi Contributions to Understanding Vector-Pathogen Interactions
7. RNAi: From Basic Research to Control Strategies
7.1. RNAi Contributes to the Identification of Candidate Transgenes
| Vector Species | Transgene | Pathogen(s) targeted | Reference for RNAi of the transgene |
|---|---|---|---|
| Mosquito immune response genes | |||
| Ae. aegypti | Defensin A | Micrococcus luteus | [231,232,233,234,235,236] |
| Plasmodium gallinaceum | |||
| Enterobacter cloacae | |||
| Ae. aegypti | Cecropin A | Enterobacter cloacae | |
| An. gambiae | Plasmodium berghei | ||
| Ae. aegypti | REL-genes | Plasmodium spp. | |
| Bacillus subtilis | |||
| (REL1 and REL2) | |||
| Escherichia coli | |||
| Others | |||
| An. stephensi | SM1 | Plasmodium berghei | [237] |
| Ae. aegypti | 30Ka; 30Kb | Dengue Virus | [238] |
| Ae. aegypti | Anti-DENV2 | Dengue Virus | [179,184] |
7.2. Paratransgenesis
7.3. RNAi Together with Integrated High-Throughput Technologies Pave the Way for the Future in Combatting Pathogen and Their Vectors
Acknowledgements
References
- Kolev, N.G.; Tschudi, C.; Ullu, E. RNA interference in protozoan parasites: achievements and challenges. Eukaryot Cell. 2011, 10, 1156–1163. [Google Scholar] [CrossRef]
- Sutherst, R.W. Global change and human vulnerability to vector-borne diseases. Clin. Microbiol. Rev. 2004, 17, 136–173. [Google Scholar]
- Rogers, D.J.; Randolph, S.E. Climate change and vector-borne diseases. Adv. Parasitol. 2006, 62, 345–381. [Google Scholar] [CrossRef]
- Severson, D.W.; Behura, S.K. Mosquito genomics: progress and challenges. Annu. Rev. Entomol. 2012, 57, 143–166. [Google Scholar]
- Gadelha, C.; Holden, J.M.; Allison, H.C.; Field, M.C. Specializations in a successful parasite: what makes the bloodstream-form African trypanosome so deadly? Mol. Biochem. Parasitol. 2011, 179, 51–58. [Google Scholar] [CrossRef]
- Suarez, C.E.; Noh, S. Emerging perspectives in the research of bovine babesiosis and anaplasmosis. Vet. Parasitol. 2011, 180, 109–125. [Google Scholar] [CrossRef]
- Schuijt, T.; Hovius, J.W.; van der Poll, T.; van Dam, A.P.; Fikrig, E. Lyme borreliosis vaccination: the facts, the challenge, the future. Trends. Parasitol. 2011, 40–47. [Google Scholar]
- Mito, T.; Nakamura, T.; Bando, T.; Hideyo, O.; Sumihare, N. The advent of RNA interference in entomology. Entomol. Sci. 2011, 14, 1–8. [Google Scholar]
- Lawson, D.; Arensburger, P.; Atkinson, P.; Besansky, N.J.; Bruggner, R.V.; Butler, R.; Campbell, K.S.; Christophides, G.K.; Christley, S.; Dialynas, E.; et al. VectorBase: A home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2007, 35, D503–D505. [Google Scholar] [CrossRef]
- Topalis, P.; Dialynas, E.; Mitraka, E.; Deligianni, E.; Siden-Kiamos, I.; Louis, C. A set of ontologies to drive tools for the control of vector-borne diseases. J. Biomed. Inform. 2011, 44, 42–47. [Google Scholar]
- Mills, J.N.; Gage, K.L.; Khan, A.S. Potential influence of climate change on vector-borne and zoonotic diseases: a review and proposed research plan. Environ. Health Perspect 2010, 118, 1507–1514. [Google Scholar] [CrossRef]
- Lawson, D.; Arensburger, P.; Atkinson, P.; Besansky, N.J.; Bruggner, R.V.; Butler, R.; Campbell, K.S.; Christophides, G.K.; Christley, S.; Dialynas, E.; et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009, 37, D583–D587. [Google Scholar] [CrossRef]
- Liu, Q.; Paroo, Z. Biochemical principles of small RNA pathways. Annu Rev. Biochem. 2010, 79, 295–319. [Google Scholar]
- Hammond, S.M. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005, 579, 5822–5829. [Google Scholar] [CrossRef]
- Walshe, D.P.; Lehane, S.M.; Lehane, M.J.; Haines, L.R. Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol. Biol. 2009, 18, 11–19. [Google Scholar] [CrossRef]
- Benoit, J.B.; Yang, G.; Krause, T.B.; Patrick, K.R.; Aksoy, S.; Attardo, G.M. Lipophorin acts as a shuttle of lipids to the milk gland during tsetse fly pregnancy. J. Insect Physiol. 2011, 57, 1553–1561. [Google Scholar]
- Caljon, G.; De Ridder, K.; De Baetselier, P.; Coosemans, M.; Van Den Abbeele, J. Identification of a tsetse fly salivary protein with dual inhibitory action on human platelet aggregation. PLoS One 2010, 5, e9671. [Google Scholar]
- Durand-Dubief, M.; Bastin, P. TbAGO1, an argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 2003, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Djikeng, A.; Tschudi, C.; Ullu, E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol. Cell Biol. 2004, 24, 420–427. [Google Scholar] [CrossRef]
- Shi, H.; Tschudi, C.; Ullu, E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA 2006, 12, 2063–2072. [Google Scholar] [CrossRef]
- Patrick, K.L.; Shi, H.; Kolev, N.G.; Ersfeld, K.; Tschudi, C.; Ullu, E. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 2009, 106, 17933–17938. [Google Scholar]
- Barnes, R.L.; Shi, H.; Kolev, N.G.; Tschudi, C.; Ullu, E. Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery. PLoS Pathog 2012, 8, e1002678. [Google Scholar] [CrossRef]
- Araujo, R.N.; Santos, A.; Pinto, F.S.; Gontijo, N.F.; Lehane, M.J.; Pereira, M.H. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem. Mol. Biol. 2006, 36, 683–693. [Google Scholar]
- Paim, R.M.; Araujo, R.N.; Soares, A.C.; Lemos, L.C.; Tanaka, A.S.; Gontijo, N.F.; Lehane, M.J.; Pereira, M.H. Influence of the intestinal anticoagulant in the feeding performance of triatomine bugs (Hemiptera; Reduviidae). Int. J. Parasitol. 2011, 41, 765–773. [Google Scholar] [CrossRef]
- Garcia Silva, M.R.; Tosar, J.P.; Frugier, M.; Pantano, S.; Bonilla, B.; Esteban, L.; Serra, E.; Rovira, C.; Robello, C.; Cayota, A. Cloning, characterization and subcellular localization of a Trypanosoma cruzi argonaute protein defining a new subfamily distinctive of trypanosomatids. Gene 2010, 15, 26–35. [Google Scholar]
- Kurscheid, S.; Lew-Tabor, A.E.; Rodriguez Valle, M.; Bruyeres, A.G.; Doogan, V.J.; Munderloh, U.G.; Guerrero, F.D.; Barrero, R.A.; Bellgard, M.I. Evidence of a tick RNAi pathway by comparative genomics and reverse genetics screen of targets with known loss-of-function phenotypes in Drosophila. BMC Mol. Biol. 2009, 10, 26. [Google Scholar] [CrossRef]
- Campbell, C.L.; Black, W.C. t.; Hess, A.M.; Foy, B.D. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics 2008, 9, 425. [Google Scholar]
- Xue, X.; Zhang, Q.; Huang, Y.; Feng, L.; Pan, W. No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar J. 2008, 7, 47. [Google Scholar] [CrossRef]
- Baum, J.; Papenfuss, A.T.; Mair, G.R.; Janse, C.J.; Vlachou, D.; Waters, A.P.; Cowman, A.F.; Crabb, B.S.; de Koning-Ward, T.F. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids. Res. 2009, 37, 3788–3798. [Google Scholar]
- Tomari, Y.; Zamore, P.D. Perspective: machines for RNAi. Genes Dev. 2005, 19, 517–529. [Google Scholar] [CrossRef]
- Sashital, D.; Doudna, J.A. Structural Insights into RNA Interference. Curr. Opin. Struct. Biol. 2010, 20, 90–97. [Google Scholar]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol 2010, 56, 227–235. [Google Scholar] [CrossRef]
- Winston, W.M.; Molodowitch, C.; Hunter, C.P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 2002, 295, 2456–2459. [Google Scholar] [CrossRef]
- Wu-yang, Z.; Guo-dong, L. Research on basis of reverse genetics system of a Sindbis-like virus XJ-160. Virol. J. 2011, 8, 519. [Google Scholar]
- Jose, A.M.; Smith, J.J.; Hunter, C.P. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc. Natl. Acad. Sci. USA 2009, 106, 2283–2288. [Google Scholar] [CrossRef]
- Calixto, A.; Chelur, D.; Topalidou, I.; Chen, X.; Chalfie, M. Enhanced neuronal RNAi in C. elegans using SID-1. Nature Meth. 2010, 7, 554–559. [Google Scholar] [CrossRef]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossman, D.; Bucher, G. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008, 9, R10. [Google Scholar]
- Ulvila, J.; Parikka, M.; Kleino, A.; Sormunen, R.; Ezekowitz, R.A.; Kocks, C.; Ramet, M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J. Biol. Chem. 2006, 281, 14370–14375. [Google Scholar]
- Saleh, M.C.; van Rij, R.P.; Hekele, A.; Gillis, A.; Foley, E.; O'Farrell, P.H.; Andino, R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell. Biol. 2006, 8, 793–802. [Google Scholar] [CrossRef]
- McManus, M.T.; Sharp, P.A. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002, 3, 737–747. [Google Scholar]
- Agrawal, N.; Dasaradhi, P.V.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001, 15, 188–200. [Google Scholar]
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000, 101, 25–33. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar]
- Bernstein, E.; Denli, A.M.; Hannon, G.J. The rest is silence. RNA 2001, 7, 1509–1521. [Google Scholar]
- Zhang, J.; Hua, Z.C. Targeted gene silencing by small interfering RNA-based knock-down technology. Curr. Pharm. Biotechnol. 2004, 5, 1–7. [Google Scholar]
- Yan, K.S.; Yan, S.; Farooq, A.; Han, A.; Zeng, L.; Zhou, M.M. Structure and conserved RNA binding of the PAZ domain. Nature 2003, 426, 468–474. [Google Scholar]
- Song, J.J.; Liu, J.; Tolia, N.H.; Schneiderman, J.; Smith, S.K.; Martienssen, R.A.; Hannon, G.J.; Joshua-Tor, L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003, 10, 1026–1032. [Google Scholar] [CrossRef]
- Ma, J.B.; Ye, K.; Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 2004, 429, 318–322. [Google Scholar]
- Zhang, H.; Kolb, F.A.; Brondani, V.; Billy, E.; Filipowicz, W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002, 21, 5875–5885. [Google Scholar] [CrossRef]
- Macrae, I.J.; Li, F.; Zhou, K.; Cande, W.Z.; Doudna, J.A. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 73–80. [Google Scholar]
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef]
- MacRae, I.J.; Zhou, K.; Doudna, J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007, 14, 934–940. [Google Scholar]
- Lau, P.; Potter, C.S.; Carragher, B.; MacRae2, I.J. Structure of the Human Dicer-TRBP Complex by Electron Microscopy. Structure 2009, 14, 1326–1332. [Google Scholar]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef]
- Ma, E.; MacRae, I.J.; Kirsch, J.F.; Doudna, J.A. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008, 380, 237–243. [Google Scholar]
- Dlakic, M. DUF283 domain of Dicer proteins has a double-stranded RNA-binding fold. Bioinformatics 2006, 22, 2711–2714. [Google Scholar] [CrossRef]
- Sasaki, T.; Shimizu, N. Evolutionary conservation of a unique amino acid sequence in human DICER protein essential for binding to Argonaute family proteins. Gene 2007, 396, 312–320. [Google Scholar]
- Provost, P.; Dishart, D.; Doucet, J.; Frendewey, D.; Samuelsson, B.; Radmark, O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002, 21, 5864–5874. [Google Scholar] [CrossRef]
- Kawamata, T.; Tomari, Y. Making RISC. Trends Biochem Sci 2010, 35, 368–376. [Google Scholar]
- Jaskiewicz, L.; Filipowicz, W. Role of Dicer in posttranscriptional RNA silencing. Curr. Top. Microbiol. Immunol. 2008, 320, 77–97. [Google Scholar] [CrossRef]
- Wang, H.; Noland, C.; Siridechadilok, B.; Taylor, D.W.; Ma, E.; Felderer, K.; Doudna, J.A.; Nogales, E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009, 16, 1148–1153. [Google Scholar]
- Tomari, Y.; Du, T.; Haley, B.; Schwarz, D.S.; Bennett, R.; Cook, H.A.; Koppetsch, B.S.; Theurkauf, W.E.; Zamore, P.D. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 2004, 116, 831–841. [Google Scholar] [CrossRef]
- Tomari, Y.; Matranga, C.; Haley, B.; Martinez, N.; Zamore, P.D. A protein sensor for siRNA asymmetry. Science 2004, 306, 1377–1380. [Google Scholar]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar]
- Pham, J.W.; Pellino, J.L.; Lee, Y.S.; Carthew, R.W.; Sontheimer, E.J. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 2004, 117, 83–94. [Google Scholar] [CrossRef]
- Pham, J.W.; Sontheimer, E.J. The Making of an siRNA. Mol. Cell 2004, 15, 163–164. [Google Scholar]
- Rand, T.A.; Petersen, S.; Du, F.; Wang, X. Argonaute2 Cleaves the Anti-Guide Strand of siRNA during RISC Activation. Cell 2005, 123, 621–629. [Google Scholar] [CrossRef]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef]
- van den Berg, A.; Mols, J.; Han, J. RISC-Target Interaction: Cleavage and Translational Suppression. Biochimica et Biophysica 2008, 1779, 668–677. [Google Scholar]
- Hutvagner, G.; Simar, M.J. Argonaute proteins: key player in RNA silencing. Nat. Rev. Mol. Biol. 2008, 9, 22–31. [Google Scholar]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef]
- Song, J.J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of Argonuate and its implication for RISC slicer activity. Science 2004, 305, 1434–1437. [Google Scholar]
- Sasaki, T.; Shiohama, A.; Minoshima, S.; Shimizu, N. Identification of eight members of the Argonaute family in the human genome. Genomics 2003, 82, 323–330. [Google Scholar]
- Yigit, E.; Batista, P.J.; Bei, Y.; Pang, K.M.; Chen, C.C.; Tolia, N.H.; Joshua-Tor, L.; Mitani, S.; Simard, M.J.; Mello, C.C. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 2006, 127, 747–757. [Google Scholar] [CrossRef]
- Caudy, A.A.; Ketting, R.F.; Hammond, S.M.; Denli, A.M.; Bathoorn, A.M. P.; Tops, B.B. J.; Silva, J.M.; Myers, M.M.; Hannon, G.J.; Plasterk, R.H. A. A micrococcal nuclease homologue in RNAi effector complexes. Nature 2003, 425, 411–414. [Google Scholar]
- Caudy, A.A.; Myers, M.; Hannon, G.J.; Hammond, S.M. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002, 16, 2491–2496. [Google Scholar] [CrossRef]
- Hall, T.M. Structure and function of argonaute proteins. Structure 2005, 13, 1403–1408. [Google Scholar]
- Ma, J.B.; Yuan, Y.R.; Meister, G.; Pei, Y.; Tuschl, T.; Patel, D.J. Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005, 434, 666–670. [Google Scholar]
- Parker, J.S.; Roe, S.M.; Barford, D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004, 23, 4727–4737. [Google Scholar] [CrossRef]
- Nykanen, A.; Haley, B.; Zamore, P.D. ATP Requirements and Small Interfering RNA Structure in the RNA Interference Pathway. Cell 2001, 107, 309–321. [Google Scholar]
- Haley, B.; Zamore, P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004, 11, 599–606. [Google Scholar] [CrossRef]
- Zamore, P.D.; Haley, B. Ribo-gnome: The big world of small RNAs. Science 2005, 309, 1519–1524. [Google Scholar]
- Cerutti, H. RNA interference: Traveling in the cell and gaining functions? Trends Genet. 2003, 19, 39–46. [Google Scholar] [CrossRef]
- Karim, S.; Ramakrishnan, V.J.; Tucker, J.S.; Essenberg, R.C.; Sauer, J.R. Amblyomma americanum salivary glands:double-stranded RNA-mediated gene silencing of synaptobrevin homologue and inhibition of PGE2 stimulated protein secretion. Insect Biochem. Mol. Biol. 2004, 34, 407–413. [Google Scholar] [CrossRef]
- Nijhof, A.M.; Taoufik, A.; de la Fuente, J.; Kocan, K.M.; de Vries, E.; Jongejan, F. Gene silencing of the tick protective antigens, Bm86, Bm91 and subolesin, in the one-host tick Boophilus microplus by RNA interference. Int. J. Parasitol. 2007, 37, 653–662. [Google Scholar] [CrossRef]
- Soares, C.A.; Lima, C.M.; Dolan, M.C.; Piesman, J.; Beard, C.B.; Zeidner, N.S. Capillary feeding of specific dsRNA induces silencing of the isac gene in nymphal Ixodes scapularis ticks. Insect Mol. Biol. 2005, 14, 443–452. [Google Scholar] [CrossRef]
- Barnard, A.-C.; Nijhof, A.M.; Gaspar, A.R.; Neitz, A.W.; Jongejan, F.; Maritz-Olivier, C. Expression profiling, gene silencing and transcriptional networking of metzincin metalloproteases in the cattle tick, Rhipicephalus (Boophilus) microplus. Vet. Parasitol. 2012, 186, 403–414. [Google Scholar] [CrossRef]
- Tijsterman, M.; May, R.C.; Simmer, F.; Okihara, K.L.; Plasterk, R.H. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr. Biol. 2004, 14, 111–116. [Google Scholar] [CrossRef]
- Sijen, T.; Fleenor, J.; Simmer, F.; Thijssen, K.L.; Parrish, S.; Timmons, L.; Plasterk, R.H.A.; Fire, A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 2001, 107, 465–476. [Google Scholar]
- Aung, K.M.; Boldbaatar, D.; Umemiya-Shirafuji, R.; Liao, M.; Xuenan, X.; Suzuki, H.; Galay, R.L.; Tanaka, T.; Fujisaki, K. Scavenger receptor mediates systemic RNA interference in ticks. PLoS One 2011, 6, e28407. [Google Scholar]
- Smardon, A.; Spoerke, J.M.; Stacey, S.C.; Klein, M.E.; Mackin, N.; Maine, E.M. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000, 10, 169–178. [Google Scholar] [CrossRef]
- Buchon, N.; Vaury, C. RNAi: A defensive RNA-silencing against viruses and transposable elements. Heredity 2006, 96, 196–202. [Google Scholar]
- Jongejan, F.; Uilenberg, G. The global importance of ticks. Parasitology 2004, 129, S3–S14. [Google Scholar] [CrossRef]
- Guerrero, F.D.; Nene, V.M.; George, J.E.; Barker, S.C.; Willadsen, P. Sequencing a new target genome: The Boophilus microplus (Acari: Ixodidae) genome project. J. Med. Entomol. 2006, 43, 9–16. [Google Scholar] [CrossRef]
- Bellgard, M.I.; Moolhuijzen, P.M.; Guerrero, F.D.; Schibeci, D.; Rodriguez-Valle, M.; Peterson, D.G.; Dowd, S.E.; Barrero, R.; Hunter, A.; Miller, R.J.; et al. CattleTickBase: An integrated Internet-based bioinformatics resource for Rhipicephalus (Boophilus) microplus. Int. J. Parasitol. 2012, 42, 161–169. [Google Scholar] [CrossRef]
- Pagel Van Zee, J.; Geraci, N.S.; Guerrero, F.D.; Wikel, S.K.; Stuart, J.J.; Nene, V.M.; Hill, C.A. Tick genomics: the Ixodes genome project and beyond. Int. J. Parasitol. 2007, 37, 1297–1305. [Google Scholar] [CrossRef]
- Aljamali, M.N.; Sauer, J.R.; Essenberg, R.C. RNA interference: applicability in tick research. Exp. Appl. Acarol. 2002, 28, 89–96. [Google Scholar] [CrossRef]
- de la Fuente, J.; Almazan, C.; Blas-Machado, U.; Naranjo, V.; Mangold, A.J.; Blouin, E.F.; Gortazar, C.; Kocan, K.M. The tick protective antigen, 4D8, is a conserved protein involved in modulation of tick blood ingestion and reproduction. Vaccine 2006, 24, 4082–4095. [Google Scholar] [CrossRef]
- Miyoshi, T.; Tsuji, N.; Islam, M.K.; Kamio, T.; Fujisaki, K. Gene silencing of a cubilin-related serine proteinase from the hard tick Haemaphysalis longicornis by RNA interference. J. Vet. Med. Sci. 2004, 66, 1471–1473. [Google Scholar] [CrossRef]
- Manzano-Roman, R.; Diaz-Martin, V.; Oleaga, A.; Siles-Lucas, M.; Perez-Sanchez, R. Subolesin/akirin orthologs from Ornithodoros spp. soft ticks: Cloning, RNAi gene silencing and protective effect of the recombinant proteins. Vet. Parasitol. 2012, 185, 248–259. [Google Scholar] [CrossRef]
- Grbic, M.; Van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouze, P.; Grbic, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.; Ortego, F.; et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar]
- Campbell, E.M.; Budge, G.E.; Bowman, A.S. Gene-knockdown in the honey bee mite Varroa destructor by a non-invasive approach: studies on a glutathione S-transferase. Parasit Vectors 2010, 3, 73. [Google Scholar] [CrossRef] [Green Version]
- Khila, A.; Grbic, M. Gene silencing in the spider mite Tetranychus urticae: dsRNA and siRNA parental silencing of the Distal-less gene. Dev. Genes Evol. 2007, 217, 241–251. [Google Scholar] [CrossRef]
- Ceraul, S.M.; Chung, A.; Sears, K.T.; Popov, V.L.; Beier-Sexton, M.; Rahman, M.S.; Azad, A.F. A Kunitz protease inhibitor from Dermacentor variabilis, a vector for spotted fever group rickettsiae, limits Rickettsia montanensis invasion. Infect. Immun. 2011, 79, 321–329. [Google Scholar] [CrossRef]
- Narasimhan, S.; Sukumaran, B.; Bozdogan, U.; Thomas, V.; Liang, X.; DePonte, K.; Marcantonio, N.; Koski, R.A.; Anderson, J.F.; Kantor, F.; et al. A tick antioxidant facilitates the Lyme disease agent's successful migration from the mammalian host to the arthropod vector. Cell Host Microbe 2007, 2, 7–18. [Google Scholar]
- Pedra, J.H.; Narasimhan, S.; Rendic, D.; DePonte, K.; Bell-Sakyi, L.; Wilson, I.B.; Fikrig, E. Fucosylation enhances colonization of ticks by Anaplasma phagocytophilum. Cell Microbiol. 2010, 12, 1222–1234. [Google Scholar] [CrossRef]
- Seinen, E.; Burgerhof, J.G.; Jansen, R.C.; Sibon, O.C. RNAi experiments in D. melanogaster: solutions to the overlooked problem of off-targets shared by independent dsRNAs. PLoS One 2010, 5, e13119. [Google Scholar]
- Sigoillot, F.D.; Lyman, S.; Huckins, J.F.; Adamson, B.; Chung, E.; Quattrochi, B.; King, R.W. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Meth. 2012, 9, 363–366. [Google Scholar]
- Lew-Tabor, A.E.; Kurscheid, S.; Barrero, R.; Gondro, C.; Moolhuijzen, P.M.; Rodriguez Valle, M.; Morgan, J.A.; Covacin, C.; Bellgard, M.I. Gene expression evidence for off-target effects caused by RNA interference-mediated gene silencing of Ubiquitin-63E in the cattle tick Rhipicephalus microplus. Int. J. Parasitol. 2011, 41, 1001–1014. [Google Scholar] [CrossRef]
- Campbell, E.M.; Burdin, M.; Hoppler, S.; Bowman, A.S. Role of an aquaporin in the sheep tick Ixodes ricinus: assessment as a potential control target. Int. J. Parasitol. 2010, 40, 15–23. [Google Scholar] [CrossRef]
- Buresova, V.; Hajdusek, O.; Franta, Z.; Sojka, D.; Kopacek, P. IrAM-An alpha2-macroglobulin from the hard tick Ixodes ricinus: characterization and function in phagocytosis of a potential pathogen Chryseobacterium indologenes. Dev. Comp. Immunol. 2009, 33, 489–498. [Google Scholar] [CrossRef]
- Narasimhan, S.; Montgomery, R.R.; DePonte, K.; Tschudi, C.; Marcantonio, N.; Anderson, J.F.; Sauer, J.R.; Cappello, M.; Kantor, F.S.; Fikrig, E. Disruption of Ixodes scapularis anticoagulation by using RNA interference. Proc. Natl. Acad. Sci. USA 2004, 101, 1141–1146. [Google Scholar]
- Umemiya-Shirafuji, R.; Tanaka, T.; Boldbaatar, D.; Fujisaki, K. Akt is an essential player in regulating cell/organ growth at the adult stage in the hard tick Haemaphysalis longicornis. Insect Biochem. Mol. Biol. 2012, 42, 164–173. [Google Scholar] [CrossRef]
- Alim, M.A.; Tsuji, N.; Miyoshi, T.; Islam, M.K.; Hatta, T.; Fujisaki, K. Legumains from the hard tick Haemaphysalis longicornis play modulatory roles in blood feeding and gut cellular remodelling and impact on embryogenesis. Int. J. Parasitol. 2009, 39, 97–107. [Google Scholar] [CrossRef]
- Kocan, K.M.; Manzano-Roman, R.; de la Fuente, J. Transovarial silencing of the subolesin gene in three-host ixodid tick species after injection of replete females with subolesin dsRNA. Parasitol. Res. 2007, 100, 1411–1415. [Google Scholar]
- Merino, O.; Almazan, C.; Canales, M.; Villar, M.; Moreno-Cid, J.A.; Estrada-Pena, A.; Kocan, K.M.; de la Fuente, J. Control of Rhipicephalus (Boophilus) microplus infestations by the combination of subolesin vaccination and tick autocidal control after subolesin gene knockdown in ticks fed on cattle. Vaccine 2011, 29, 2248–2254. [Google Scholar] [CrossRef]
- Garcia, S.; Billecocq, A.; Crance, J.M.; Munderloh, U.; Garin, D.; Bouloy, M. Nairovirus RNA sequences expressed by a Semliki Forest virus replicon induce RNA interference in tick cells. J. Virol. 2005, 79, 8942–8947. [Google Scholar]
- Barry, G.; Alberdi, P.; Schnettler, E.; Weisheit, S.; Kohl, A.; Fazakerley, J.K.; Bell-Sakyi, L. Gene silencing in tick cell lines using small interfering or long double-stranded RNA. Exp. Appl. Acarol. 2012, in press.. [Google Scholar]
- Karim, S.; Kenny, B.; Troiano, E.; Mather, T.N. RNAi-mediated gene silencing in tick synganglia: a proof of concept study. BMC Biotechnol. 2008, 8, 30. [Google Scholar]
- Aljamali, M.N.; Bior, A.D.; Sauer, J.R.; Essenberg, R.C. RNA interference in ticks: a study using histamine binding protein dsRNA in the female tick Amblyomma americanum. Insect Mol. Biol. 2003, 12, 299–305. [Google Scholar] [CrossRef]
- Gong, H.; Umemiya, R.; Zhou, J.; Liao, M.; Zhang, H.; Jia, H.; Nishikawa, Y.; Xuan, X.; Fujisaki, K. Blocking the secretion of saliva by silencing the HlYkt6 gene in the tick Haemaphysalis longicornis. Insect Biochem. Mol. Biol. 2009, 39, 372–381. [Google Scholar] [CrossRef]
- Karim, S.; Troiano, E.; Mather, T.N. Functional genomics tool: gene silencing in Ixodes scapularis eggs and nymphs by electroporated dsRNA. BMC Biotechnol. 2010, 10, 1. [Google Scholar] [CrossRef]
- Beaufays, J.; Adam, B.; Menten-Dedoyart, C.; Fievez, L.; Grosjean, A.; Decrem, Y.; Prevot, P.P.; Santini, S.; Brasseur, R.; Brossard, M.; et al. Ir-LBP, an Ixodes ricinus tick salivary LTB4-binding lipocalin, interferes with host neutrophil function. PLoS One 2008, 3, e3987. [Google Scholar]
- Dai, J.; Narasimhan, S.; Zhang, L.; Liu, L.; Wang, P.; Fikrig, E. Tick histamine release factor is critical for Ixodes scapularis engorgement and transmission of the lyme disease agent. PLoS Pathog. 2010, 6, e1001205. [Google Scholar] [CrossRef]
- Decrem, Y.; Mariller, M.; Lahaye, K.; Blasioli, V.; Beaufays, J.; Zouaoui Boudjeltia, K.; Vanhaeverbeek, M.; Cerutti, M.; Brossard, M.; Vanhamme, L.; et al. The impact of gene knock-down and vaccination against salivary metalloproteases on blood feeding and egg laying by Ixodes ricinus. Int. J. Parasitol. 2008, 38, 549–560. [Google Scholar] [CrossRef]
- Gao, X.; Shi, L.; Zhou, Y.; Cao, J.; Zhang, H.; Zhou, J. Characterization of the anticoagulant protein Rhipilin-1 from the Rhipicephalus haemaphysaloides tick. J. Insect Physiol. 2011, 57, 339–343. [Google Scholar] [CrossRef]
- Guo, X.; Booth, C.J.; Paley, M.A.; Wang, X.; DePonte, K.; Fikrig, E.; Narasimhan, S.; Montgomery, R.R. Inhibition of neutrophil function by two tick salivary proteins. Infect. Immun. 2009, 77, 2320–2329. [Google Scholar]
- Karim, S.; Ramakrishnan, V.G.; Tucker, J.S.; Essenberg, R.C.; Sauer, J.R. Amblyomma americanum salivary gland homolog of nSec1 is essential for saliva protein secretion. Biochem. Biophys Res. Commun. 2004, 324, 1256–1263. [Google Scholar] [CrossRef]
- Franta, Z.; Sojka, D.; Frantova, H.; Dvorak, J.; Horn, M.; Srba, J.; Talacko, P.; Mares, M.; Schneider, E.; Craik, C.S.; et al. IrCL1 - the haemoglobinolytic cathepsin L of the hard tick, Ixodes ricinus. Int. J. Parasitol. 2011, 41, 1253–1262. [Google Scholar] [CrossRef]
- Miyoshi, T.; Tsuji, N.; Islam, M.K.; Alim, M.A.; Hatta, T.; Huang, X.; Fujisaki, K. A set of serine proteinase paralogs are required for blood-digestion in the ixodid tick Haemaphysalis longicornis. Parasitol. Int. 2008, 57, 499–505. [Google Scholar] [CrossRef]
- Hajdusek, O.; Sojka, D.; Kopacek, P.; Buresova, V.; Franta, Z.; Sauman, I.; Winzerling, J.; Grubhoffer, L. Knockdown of proteins involved in iron metabolism limits tick reproduction and development. Proc. Natl. Acad. Sci. USA 2009, 106, 1033–1038. [Google Scholar]
- Guo, X.; Reuben Kaufman, W. Identification of two genes essential for sperm development in the male tick Amblyomma hebraeum Koch (Acari: Ixodidae). Insect Biochem. Mol. Biol. 2008, 38, 721–729. [Google Scholar] [CrossRef]
- Smith, A.; Guo, X.; de la Fuente, J.; Naranjo, V.; Kocan, K.M.; Kaufman, W.R. The impact of RNA interference of the subolesin and voraxin genes in male Amblyomma hebraeum (Acari: Ixodidae) on female engorgement and oviposition. Exp. Appl. Acarol. 2009, 47, 71–86. [Google Scholar] [CrossRef]
- Buresova, V.; Hajdusek, O.; Franta, Z.; Loosova, G.; Grunclova, L.; Levashina, E.A.; Kopacek, P. Functional genomics of tick thioester-containing proteins reveal the ancient origin of the complement system. J. Innate Immun. 2011, 3, 623–630. [Google Scholar]
- de la Fuente, J.; Almazan, C.; Blouin, E.F.; Naranjo, V.; Kocan, K.M. RNA interference screening in ticks for identification of protective antigens. Parasitol. Res. 2005, 96, 137–141. [Google Scholar] [CrossRef]
- de la Fuente, J.; Manzano-Roman, R.; Naranjo, V.; Kocan, K.M.; Zivkovic, Z.; Blouin, E.F.; Canales, M.; Almazan, C.; Galindo, R.C.; Step, D.L.; et al. Identification of protective antigens by RNA interference for control of the lone star tick, Amblyomma americanum. Vaccine 2010, 28, 1786–1795. [Google Scholar]
- de la Fuente, J.; Moreno-Cid, J.A.; Canales, M.; Villar, M.; de la Lastra, J.M.; Kocan, K.M.; Galindo, R.C.; Almazan, C.; Blouin, E.F. Targeting arthropod subolesin/akirin for the development of a universal vaccine for control of vector infestations and pathogen transmission. Vet. Parasitol. 2011, 181, 17–22. [Google Scholar]
- de la Fuente, J.; Almazan, C.; Naranjo, V.; Blouin, E.F.; Meyer, J.M.; Kocan, K.M. Autocidal control of ticks by silencing of a single gene by RNA interference. Biochem. Biophys Res. Commun. 2006, 344, 332–338. [Google Scholar] [CrossRef]
- de la Fuente, J.; Blouin, E.F.; Manzano-Roman, R.; Naranjo, V.; Almazan, C.; Perez de la Lastra, J.M.; Zivkovic, Z.; Jongejan, F.; Kocan, K.M. Functional genomic studies of tick cells in response to infection with the cattle pathogen, Anaplasma marginale. Genomics 2007, 90, 712–722. [Google Scholar] [CrossRef]
- Rachinsky, A.; Guerrero, F.D.; Scoles, G.A. Differential protein expression in ovaries of uninfected and Babesia-infected southern cattle ticks, Rhipicephalus (Boophilus) microplus. Insect Biochem. Mol. Biol. 2007, 37, 1291–1308. [Google Scholar] [CrossRef]
- Sukumaran, B.; Narasimhan, S.; Anderson, J.F.; DePonte, K.; Marcantonio, N.; Krishnan, M.N.; Fish, D.; Telford, S.R.; Kantor, F.S.; Fikrig, E. An Ixodes scapularis protein required for survival of Anaplasma phagocytophilum in tick salivary glands. J. Exp. Med. 2006, 203, 1507–1517. [Google Scholar] [CrossRef]
- Heekin, A.M.; Guerrero, F.D.; Bendele, K.G.; Saldivar, L.; Scoles, G.A.; Gondro, C.; Nene, V.; Djikeng, A.; Brayton, K.A. Analysis of Babesia bovis infection-induced gene expression changes in larvae from the cattle tick, Rhipicephalus (Boophilus) microplus. Parasit Vectors 2012, 5, 162. [Google Scholar] [CrossRef]
- Das, S.; Banerjee, G.; DePonte, K.; Marcantonio, N.; Kantor, F.S.; Fikrig, E. Salp25D, an Ixodes scapularis antioxidant, is 1 of 14 immunodominant antigens in engorged tick salivary glands. J. Infect. Dis 2001, 184, 1056–1064. [Google Scholar] [CrossRef]
- Ramamoorthi, N.; Narasimhan, S.; Pal, U.; Bao, F.; Yang, X.F.; Fish, D.; Anguita, J.; Norgard, M.V.; Kantor, F.S.; Anderson, J.F.; et al. The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature 2005, 436, 573–577. [Google Scholar]
- Hynes, W.L.; Stokes, M.M.; Hensley, S.M.; Todd, S.M.; Sonenshine, D.E. Using RNA interference to determine the role of varisin in the innate immune system of the hard tick Dermacentor variabilis (Acari: Ixodidae). Exp. Appl. Acarol. 2008, 46, 7–15. [Google Scholar] [CrossRef]
- Antunes, S.; Galindo, R.C.; Almazan, C.; Rudenko, N.; Golovchenko, M.; Grubhoffer, L.; Shkap, V.; do Rosario, V.; de la Fuente, J.; Domingos, A. Functional genomics studies of Rhipicephalus (Boophilus) annulatus ticks in response to infection with the cattle protozoan parasite, Babesia bigemina. Int J. Parasitol. 2012, 42, 187–195. [Google Scholar] [CrossRef]
- Pittendrigh, B.R.; Berenbaum, M.R.; Seufferheld, M.J.; Margam, V.M.; Strycharz, J.P.; Yoon, K.S.; Sun, W.; Reenan, R.; Lee, S.H.; Clark, J.M. Simplify, simplify: Lifestyle and compact genome of the body louse provide a unique functional genomics opportunity. Commun. Integr. Biol. 2011, 4, 188–191. [Google Scholar]
- Kirkness, E.F.; Haas, B.J.; Sun, W.; Braig, H.R.; Perotti, M.A.; Clark, J.M.; Lee, S.H.; Robertson, H.M.; Kennedy, R.C.; Elhaik, E.; et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2010, 107, 12168–12173. [Google Scholar]
- Yoon, K.S.; Strycharz, J.P.; Baek, J.H.; Sun, W.; Kim, J.H.; Kang, J.S.; Pittendrigh, B.R.; Lee, S.H.; Clark, J.M. Brief exposures of human body lice to sublethal amounts of ivermectin over-transcribes detoxification genes involved in tolerance. Insect Mol. Biol. 2011, 20, 687–699. [Google Scholar]
- Attardo, G.M.; Hansen, I.A.; Raikhel, A.S. Nutritional regulation of vitellogenesis in mosquitoes: implications for anautogeny. Insect Biochem. Mol. Biol. 2005, 35, 661–675. [Google Scholar] [CrossRef]
- Scott, T.W.; Takken, W. Feeding strategies of anthropophilic mosquitoes result in increased risk of pathogen transmission. Trends Parasitol. 2012, 28, 114–121. [Google Scholar]
- Manzano-Roman, R.; Oleaga, A.; Perez-Sanchez, R.; Siles-Lucas, M.; Rollinson, D.; Hay, S.I. Gene Silencing in Parasites: Current Status and Future Prospects. Adv. Parasitol. 2012, 78, 1–55. [Google Scholar] [CrossRef]
- Belles, X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol. 2010, 55, 111–128. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Zhu, K.Y. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 2010, 19, 683–693. [Google Scholar] [CrossRef]
- Pridgeon, J.W.; Zhao, L.; Becnel, J.J.; Strickman, D.A.; Clark, G.G.; Linthicum, K.J. Topically applied AaeIAP1 double-stranded RNA kills female adults of Aedes aegypti. J. Med. Entomol. 2008, 45, 414–420. [Google Scholar] [CrossRef]
- Coy, M.R.; Sanscrainte, N.D.; Chalaire, K.C.; Inberg, A.; Maayan, I.; Glick, E.; Paldi, N.; Becnel, J.J. Gene silencing in adult Aedes aegypti mosquitoes through oral delivery of double-stranded RNA. J. Appl. Entomol. 2012. [Google Scholar]
- Gu, J.; Liu, M.; Deng, Y.; Peng, H.; Chen, X. Development of an efficient recombinant mosquito densovirus-mediated RNA interference system and its preliminary application in mosquito control. PLoS One 2011, 6, e21329. [Google Scholar]
- Boisson, B.; Jacques, J.C.; Choumet, V.; Martin, E.; Xu, J.; Vernick, K.; Bourgouin, C. Gene silencing in mosquito salivary glands by RNAi. FEBS Lett. 2006, 580, 1988–1992. [Google Scholar]
- Vodovar, N.; Saleh, M.-C. Of Insects and Viruses: The Role of Small RNAs in Insect Defence. Adv. Insect Physiol. 2012, 42, 1–36. [Google Scholar] [CrossRef]
- Gulia-Nuss, M.; Robertson, A.E.; Brown, M.R.; Strand, M.R. Insulin-like peptides and the target of rapamycin pathway coordinately regulate blood digestion and egg maturation in the mosquito Aedes aegypti. PLoS One 2011, 6, e20401. [Google Scholar]
- Hansen, I.A.; Attardo, G.M.; Roy, S.G.; Raikhel, A.S. Target of rapamycin-dependent activation of S6 kinase is a central step in the transduction of nutritional signals during egg development in a mosquito. J. Biol Chem 2005, 280, 20565–20572. [Google Scholar]
- Hansen, I.A.; Boudko, D.Y.; Shiao, S.H.; Voronov, D.A.; Meleshkevitch, E.A.; Drake, L.L.; Aguirre, S.E.; Fox, J.M.; Attardo, G.M.; Raikhel, A.S. AaCAT1 of the yellow fever mosquito, Aedes aegypti: a novel histidine-specific amino acid transporter from the SLC7 family. J. Biol. Chem. 2011, 286, 10803–10813. [Google Scholar]
- Roy, S.G.; Raikhel, A.S. The small GTPase Rheb is a key component linking amino acid signaling and TOR in the nutritional pathway that controls mosquito egg development. Insect Biochem. Mol. Biol. 2011, 41, 62–69. [Google Scholar]
- Roy, S.G.; Raikhel, A.S. Nutritional and hormonal regulation of the TOR effector 4E-binding protein (4E-BP) in the mosquito Aedes aegypti. FASEB J. 2012, 26, 1334–1342. [Google Scholar] [CrossRef]
- Alabaster, A.; Isoe, J.; Zhou, G.; Lee, A.; Murphy, A.; Day, W.A.; Miesfeld, R.L. Deficiencies in acetyl-CoA carboxylase and fatty acid synthase 1 differentially affect eggshell formation and blood meal digestion in Aedes aegypti. Insect Biochem. Mol. Biol. 2011, 41, 946–955. [Google Scholar] [CrossRef]
- Isoe, J.; Collins, J.; Badgandi, H.; Day, W.A.; Miesfeld, R.L. Defects in coatomer protein I (COPI) transport cause blood feeding-induced mortality in Yellow Fever mosquitoes. Proc. Natl. Acad. Sci. USA 2011, 108, E211–E217. [Google Scholar]
- Roy, S.G.; Hansen, I.A.; Raikhel, A.S. Effect of insulin and 20-hydroxyecdysone in the fat body of the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 2007, 37, 1317–1326. [Google Scholar] [CrossRef]
- Hansen, I.A.; Attardo, G.M.; Park, J.H.; Peng, Q.; Raikhel, A.S. Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proc. Natl. Acad. Sci. USA 2004, 101, 10626–10631. [Google Scholar]
- Biessmann, H.; Andronopoulou, E.; Biessmann, M.R.; Douris, V.; Dimitratos, S.D.; Eliopoulos, E.; Guerin, P.M.; Iatrou, K.; Justice, R.W.; Krober, T.; et al. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS One 2010, 5, e9471. [Google Scholar]
- Pelletier, J.; Guidolin, A.; Syed, Z.; Cornel, A.J.; Leal, W.S. Knockdown of a Mosquito Odorant-binding Protein Involved in the Sensitive Detection of Oviposition Attractants. J. Chem. Ecol. 2010, 36. [Google Scholar]
- Erdelyan, C.N.; Mahood, T.H.; Bader, T.S.; Whyard, S. Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol. Biol. 2012, 21, 119–127. [Google Scholar] [CrossRef]
- Caplen, N.J.; Zheng, Z.; Falgout, B.; Morgan, R.A. Inhibition of viral gene expression and replication in mosquito cells by dsRNA-triggered RNA interference. Mol. Ther 2002, 6, 243–251. [Google Scholar]
- Adelman, Z.N.; Sanchez-Vargas, I.; Travanty, E.A.; Carlson, J.O.; Beaty, B.J.; Blair, C.D.; Olson, K.E. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 2002, 76, 12925–12933. [Google Scholar] [CrossRef]
- Blandin, S.; Moita, L.F.; Kocher, T.; Wilm, M.; Kafatos, F.C.; Levashina, E.A. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep. 2002, 3, 852–856. [Google Scholar] [CrossRef]
- Brown, A.E.; Bugeon, L.; Crisanti, A.; Catteruccia, F. Stable and heritable gene silencing in the malaria vector Anopheles stephensi. Nucleic Acids. Res. 2003, 31, e85. [Google Scholar] [CrossRef]
- Franz, A.W.; Sanchez-Vargas, I.; Adelman, Z.N.; Blair, C.D.; Beaty, B.J.; James, A.A.; Olson, K.E. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc. Natl. Acad. Sci. USA 2006, 103, 4198–4203. [Google Scholar]
- Keene, K.M.; Foy, B.D.; Sanchez-Vargas, I.; Beaty, B.J.; Blair, C.D.; Olson, K.E. NA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2004, 101, 17240–17245. [Google Scholar]
- Blair, C.D. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011, 6, 265–277. [Google Scholar] [CrossRef]
- Attarzadeh-Yazdi, G.; Fragkoudis, R.; Chi, Y.; Siu, R.W.; Ulper, L.; Barry, G.; Rodriguez-Andres, J.; Nash, A.A.; Bouloy, M.; Merits, A.; et al. Cell-to-cell spread of the RNA interference response suppresses Semliki Forest virus (SFV) infection of mosquito cell cultures and cannot be antagonized by SFV. J. Virol. 2009, 83, 5735–5748. [Google Scholar]
- Vodovar, N.; Bronkhorst, A.W.; van Cleef, K.W.; Miesen, P.; Blanc, H.; van Rij, R.P.; Saleh, M.C. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One 2012, 7, e30861. [Google Scholar]
- Franz, A.W.; Sanchez-Vargas, I.; Piper, J.; Smith, M.R.; Khoo, C.C.; James, A.A.; Olson, K.E. Stability and loss of a virus resistance phenotype over time in transgenic mosquitoes harbouring an antiviral effector gene. Insect Mol. Biol. 2009, 18, 661–672. [Google Scholar]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef]
- Souza-Neto, J.A.; Sim, S.; Dimopoulos, G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. USA 2009, 106, 17841–17846. [Google Scholar]
- Fragkoudis, R.; Attarzadeh-Yazdi, G.; Nash, A.A.; Fazakerley, J.K.; Kohl, A. Advances in dissecting mosquito innate immune responses to arbovirus infection. J. Gen. Virol. 2009, 90, 2061–2072. [Google Scholar]
- Merkling, S.H.; van Rij, R.P. Beyond RNAi: Antiviral defense strategies in Drosophila and Mosquito. J. Insect Physiol. 2012, Jul 20, 22824741. [Google Scholar]
- Sim, S.; Dimopoulos, G. Dengue virus inhibits immune responses in Aedes aegypti cells. PLoS One 2010, 5, e10678. [Google Scholar] [CrossRef]
- Colpitts, T.M.; Cox, J.; Vanlandingham, D.L.; Feitosa, F.M.; Cheng, G.; Kurscheid, S.; Wang, P.; Krishnan, M.N.; Higgs, S.; Fikrig, E. Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 2011, 7, e1002189. [Google Scholar]
- Benedict, C.A.; Norris, P.S.; Ware, C.F. To kill or be killed: viral evasion of apoptosis. Nat. Immunol. 2002, 3, 1013–1018. [Google Scholar] [CrossRef]
- Chen, T.H.; Tang, P.; Yang, C.F.; Kao, L.H.; Lo, Y.P.; Chuang, C.K.; Shih, Y.T.; Chen, W.J. Antioxidant defense is one of the mechanisms by which mosquito cells survive dengue 2 viral infection. Virology 2011, 410, 410–417. [Google Scholar]
- Chen, T.H.; Lo, Y.P.; Yang, C.F.; Chen, W.J. Additive protection by antioxidant and apoptosis-inhibiting effects on mosquito cells with dengue 2 virus infection. PLoS Negl. Trop. Dis. 2012, 6, e1613. [Google Scholar]
- Cirimotich, C.M.; Dong, Y.; Garver, L.S.; Sim, S.; Dimopoulos, G. Mosquito immune defenses against Plasmodium infection. Dev. Comp. Immunol. 2010, 34, 387–395. [Google Scholar] [CrossRef]
- Blandin, S.A.; Levashina, E.A. Reverse genetics analysis of antiparasitic responses in the malaria vector, Anopheles gambiae. Meth. Mol. Biol. 2008, 415, 365–377. [Google Scholar]
- Marois, E. The multifaceted mosquito anti-Plasmodium response. Curr. Opin. Microbiol. 2011, 14, 429–435. [Google Scholar] [CrossRef]
- Blandin, S.; Shiao, S.H.; Moita, L.F.; Janse, C.J.; Waters, A.P.; Kafatos, F.C.; Levashina, E.A. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 2004, 116, 661–670. [Google Scholar] [CrossRef]
- Osta, M.A.; Christophides, G.K.; Kafatos, F.C. Effects of mosquito genes on Plasmodium development. Science 2004, 303, 2030–2032. [Google Scholar]
- Riehle, M.M.; Markianos, K.; Niare, O.; Xu, J.; Li, J.; Toure, A.M.; Podiougou, B.; Oduol, F.; Diawara, S.; Diallo, M.; et al. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science 2006, 312, 577–579. [Google Scholar] [CrossRef]
- Povelones, M.; Waterhouse, R.M.; Kafatos, F.C.; Christophides, G.K. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 2009, 324, 258–261. [Google Scholar]
- Fraiture, M.; Baxter, R.H.; Steinert, S.; Chelliah, Y.; Frolet, C.; Quispe-Tintaya, W.; Hoffmann, J.A.; Blandin, S.A.; Levashina, E.A. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell. Host Microbe 2009, 5, 273–284. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Devenport, M.; Jethwaney, D.; Kalume, D.E.; Pandey, A.; Anderson, V.E.; Sultan, A.A.; Kumar, N.; Jacobs-Lorena, M. Malaria parasite invasion of the mosquito salivary gland requires interaction between the Plasmodium TRAP and the Anopheles saglin proteins. PLoS Pathog. 2009, 5, e1000265. [Google Scholar] [CrossRef]
- Dong, Y.; Taylor, H.E.; Dimopoulos, G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006, 4, e229. [Google Scholar]
- Dong, Y.; Dimopoulos, G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J. Biol. Chem. 2009, 284, 9835–9844. [Google Scholar] [CrossRef]
- Shin, S.W.; Zou, Z.; Raikhel, A.S. A new factor in the Aedes aegypti immune response: CLSP2 modulates melanization. EMBO Rep. 2011, 12, 938–943. [Google Scholar] [CrossRef]
- Rodrigues, J.; Oliveira, G.A.; Kotsyfakis, M.; Dixit, R.; Molina-Cruz, A.; Jochim, R.; Barillas-Mury, C. An epithelial serine protease, AgESP, is required for Plasmodium invasion in the mosquito Anopheles gambiae. PLoS One 2012, 7, e35210. [Google Scholar]
- Chertemps, T.; Mitri, C.; Perrot, S.; Sautereau, J.; Jacques, J.C.; Thiery, I.; Bourgouin, C.; Rosinski-Chupin, I. Anopheles gambiae PRS1 modulates Plasmodium development at both midgut and salivary gland steps. PLoS One 2010, 5, e11538. [Google Scholar]
- Kajla, M.K.; Shi, L.; Li, B.; Luckhart, S.; Li, J.; Paskewitz, S.M. A new role for an old antimicrobial: lysozyme c-1 can function to protect malaria parasites in Anopheles mosquitoes. PLoS One 2011, 6, e19649. [Google Scholar]
- Mendes, A.M.; Schlegelmilch, T.; Cohuet, A.; Awono-Ambene, P.; De Iorio, M.; Fontenille, D.; Morlais, I.; Christophides, G.K.; Kafatos, F.C.; Vlachou, D. Conserved mosquito/parasite interactions affect development of Plasmodium falciparum in Africa. PLoS Pathog. 2008, 4, e1000069. [Google Scholar]
- Lavore, A.; Pagola, L.; Esponda-Behrens, N.; Rivera-Pomar, R. The gap gene giant of Rhodnius prolixus is maternally expressed and required for proper head and abdomen formation. Dev. Biol. 2012, 361, 147–155. [Google Scholar] [CrossRef]
- Araujo, R.N.; Campos, I.T.; Tanaka, A.S.; Santos, A.; Gontijo, N.F.; Lehane, M.J.; Pereira, M.H. Brasiliensin: A novel intestinal thrombin inhibitor from Triatoma brasiliensis (Hemiptera: Reduviidae) with an important role in blood intake. Int. J. Parasitol. 2007, 37, 1351–1358. [Google Scholar]
- Lye, L.F.; Owens, K.; Shi, H.; Murta, S.M.; Vieira, A.C.; Turco, S.J.; Tschudi, C.; Ullu, E.; Beverley, S.M. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010, 6, e1001161. [Google Scholar]
- Franzen, O.; Arner, E.; Ferella, M.; Nilsson, D.; Respuela, P.; Carninci, P.; Hayashizaki, Y.; Aslund, L.; Andersson, B.; Daub, C.O. The short non-coding transcriptome of the protozoan parasite Trypanosoma cruzi. PLoS Negl. Trop. Dis. 2011, 5, e1283. [Google Scholar] [CrossRef]
- Genovesio, A.; Giardini, M.A.; Kwon, Y.J.; de Macedo Dossin, F.; Choi, S.Y.; Kim, N.Y.; Kim, H.C.; Jung, S.Y.; Schenkman, S.; Almeida, I.C.; et al. Visual genome-wide RNAi screening to identify human host factors required for Trypanosoma cruzi infection. PLoS One 2011, 6, e19733. [Google Scholar]
- Taylor, M.C.; Huang, H.; Kelly, J.M. Genetic techniques in Trypanosoma cruzi. Adv. Parasitol. 2011, 75, 231–250. [Google Scholar] [CrossRef]
- Manzano-Roman, R.; Oleaga, A.; Perez-Sanchez, R.; Siles-Lucas, M. Gene silencing in parasites: current status and future prospects. Adv. Parasitol. 2012, 78, 1–55. [Google Scholar]
- Yang, G.; Attardo, G.M.; Lohs, C.; Aksoy, S. Molecular characterization of two novel milk proteins in the tsetse fly (Glossina morsitans morsitans). Insect Mol. Biol. 2010, 19, 253–262. [Google Scholar] [CrossRef]
- Lehane, M.J.; Gibson, W.; Lehane, S.M. Differential expression of fat body genes in Glossina morsitans morsitans following infection with Trypanosoma brucei brucei. Int. J. Parasitol. 2008, 38, 93–101. [Google Scholar] [CrossRef]
- Subota, I.; Rotureau, B.; Blisnick, T.; Ngwabyt, S.; Durand-Dubief, M.; Engstler, M.; Bastin, P. ALBA proteins are stage regulated during trypanosome development in the tsetse fly and participate in differentiation. Mol. Biol. Cell 2011, 22, 4205–4219. [Google Scholar]
- Haines, L.R.; Lehane, S.M.; Pearson, T.W.; Lehane, M.J. Tsetse EP protein protects the fly midgut from trypanosome establishment. PLoS Pathog. 2010, 6, e1000793. [Google Scholar] [CrossRef]
- Natesan, S.K.; Peacock, L.; Leung, K.F.; Matthews, K.R.; Gibson, W.; Field, M.C. The trypanosome Rab-related proteins RabX1 and RabX2 play no role in intracellular trafficking but may be involved in fly infectivity. PLoS One 2009, 4, e7217. [Google Scholar]
- Wang, J.; Wu, Y.; Yang, G.; Aksoy, S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. USA 2009, 106, 12133–12138. [Google Scholar] [CrossRef]
- Caljon, G.; Broos, K.; De Goeyse, I.; De Ridder, K.; Sternberg, J.M.; Coosemans, M.; De Baetselier, P.; Guisez, Y.; Den Abbeele, J.V. Identification of a functional Antigen5-related allergen in the saliva of a blood feeding insect, the tsetse fly. Insect Biochem. Mol. Biol. 2009, 39, 332–341. [Google Scholar]
- Smith, T.K.; Butikofer, P. Lipid metabolism in Trypanosoma brucei. Mol. Biochem. Parasitol. 2010, 172, 66–79. [Google Scholar] [CrossRef]
- Balana-Fouce, R.; Reguera, R.M. RNA interference in Trypanosoma brucei: a high-throughput engine for functional genomics in trypanosomatids? Trends Parasitol. 2007, 23, 348–351. [Google Scholar] [CrossRef]
- Hammarton, T.C. Cell cycle regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007, 153, 1–8. [Google Scholar] [CrossRef]
- Coutinho-Abreu, I.V.; Zhu, K.Y.; Ramalho-Ortigao, M. Transgenesis and paratransgenesis to control insect-borne diseases: current status and future challenges. Parasitol. Int. 2010, 59, 1–8. [Google Scholar]
- Malcolm, J.; Fraser, J. Insect trangenesis: Current applications and future prospects. Annu. Rev. Entomol. 2012, 57, 267–289. [Google Scholar]
- Terenius, O.; Marinotti, O.; Sieglaff, D.; James, A.A. Molecular genetic manipulation of vector mosquitoes. Cell Host Microbe 2008, 4, 417–423. [Google Scholar] [CrossRef]
- Shin, S.W.; Kokoza, V.A.; Raikhel, A.S. Transgenesis and reverse genetics of mosquito innate immunity. J. Exp. Biol. 2003, 206, 3835–3843. [Google Scholar] [CrossRef]
- Magalhaes, T.; Leandro, D.C.; Ayres, C.F. Knock-down of REL2, but not defensin A, augments Aedes aegypti susceptibility to Bacillus subtilis and Escherichia coli. Acta Trop. 2010, 113, 167–173. [Google Scholar] [CrossRef]
- Garver, L.S.; Bahia, A.C.; Das, S.; Souza-Neto, J.A.; Shiao, J.; Dong, Y.; Dimopoulos, G. Anopheles Imd pathway factors and effectors in infection intensity-dependent anti-Plasmodium action. PLoS Pathog. 2012, 8, e1002737. [Google Scholar] [CrossRef]
- Antonova, Y.; Alvarez, K.S.; Kim, Y.J.; Kokoza, V.; Raikhel, A.S. The role of NF-kappaB factor REL2 in the Aedes aegypti immune response. Insect Biochem. Mol. Biol. 2009, 39, 303–314. [Google Scholar] [CrossRef]
- Bian, G.; Shin, S.W.; Cheon, H.M.; Kokoza, V.; Raikhel, A.S. Transgenic alteration of Toll immune pathway in the female mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2005, 102, 13568–13573. [Google Scholar] [CrossRef]
- Erickson, S.M.; Xi, Z.; Mayhew, G.F.; Ramirez, J.L.; Aliota, M.T.; Christensen, B.M.; Dimopoulos, G. Mosquito infection responses to developing filarial worms. PLoS Negl. Trop. Dis. 2009, 3, e529. [Google Scholar]
- Meister, S.; Kanzok, S.M.; Zheng, X.L.; Luna, C.; Li, T.R.; Hoa, N.T.; Clayton, J.R.; White, K.P.; Kafatos, F.C.; Christophides, G.K.; et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2005 2005, 102, 11420–11425. [Google Scholar] [CrossRef]
- Dinglasan, R.R.; Kalume, D.E.; Kanzok, S.M.; Ghosh, A.K.; Muratova, O.; Pandey, A.; Jacobs-Lorena, M. Disruption of Plasmodium falciparum development by antibodies against a conserved mosquito midgut antigen. Proc. Natl. Acad. Sci. USA 2007, 104, 13461–13466. [Google Scholar]
- Mathur, G.; Sanchez-Vargas, I.; Alvarez, D.; Olson, K.E.; Marinotti, O.; James, A.A. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol. 2010, 19, 753–763. [Google Scholar] [CrossRef]
- Hurwitz, I.; Fieck, A.; Read, A.; Hillesland, H.; Klein, N.; Kang, A.; Durvasula, R. Paratransgenic control of vector borne diseases. Int. J. Biol. Sci. 2011, 7, 1334–1344. [Google Scholar]
- Cheng, Q.; Aksoy, S. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tetse flies. Insect Mol. Biol 1999, 8, 125–132. [Google Scholar]
- Favia, G.; Ricci, I.; Damiani, C.; Raddadi, N.; Crotti, E.; Marzorati, M.; Rizzi, A.; Urso, R.; Brusetti, L.; Borin, S.; et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc. Natl. Acad. Sci. USA 2007, 22, 9047–9051. [Google Scholar]
- Damiani, C.; Ricci, I.; Crotti, E.; Rossi, P.; Rizzi, A.; Scuppa, P.; Esposito, F.; Bandi, C.; Daffonchio, D.; Favia, G. Paternal transmission of symbiotic bacteria in malaria vectors. Curr. Biol. 2008, 18, R1087–R1088. [Google Scholar]
- Ren, X.; Hoiczyk, E.; Rasgon, J.L. Viral paratransgenesis in the malaria vector Anopheles gambiae. PLoS Pathog. 2008, 4, e1000135. [Google Scholar] [CrossRef]
- Ward, T.W.; Jenkins, M.S.; Afanasiev, B.N.; Edwards, M.; Duda, B.A.; Suchman, E.; Jacobs-Lorena, M.; Beaty, B.J.; Carlson, J.O. Aedes aegypti transducing densovirus pathogenesis and expression in Aedes aegypti and Anopheles gambiae larvae. Insect Mol. Biol. 2002, 10, 397–405. [Google Scholar]
- Hong-Geller, E.; Micheva-Viteva, S.N. Functional gene discovery using RNA interference-based genomic screens to combat pathogen infection. Curr. Drug Discov. Technol. 2010, 7, 86–94. [Google Scholar]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in budding yeast. Science 2009, 326, 544–550. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Barnard, A.-C.; Nijhof, A.M.; Fick, W.; Stutzer, C.; Maritz-Olivier, C. RNAi in Arthropods: Insight into the Machinery and Applications for Understanding the Pathogen-Vector Interface. Genes 2012, 3, 702-741. https://0-doi-org.brum.beds.ac.uk/10.3390/genes3040702
Barnard A-C, Nijhof AM, Fick W, Stutzer C, Maritz-Olivier C. RNAi in Arthropods: Insight into the Machinery and Applications for Understanding the Pathogen-Vector Interface. Genes. 2012; 3(4):702-741. https://0-doi-org.brum.beds.ac.uk/10.3390/genes3040702
Chicago/Turabian StyleBarnard, Annette-Christi, Ard M. Nijhof, Wilma Fick, Christian Stutzer, and Christine Maritz-Olivier. 2012. "RNAi in Arthropods: Insight into the Machinery and Applications for Understanding the Pathogen-Vector Interface" Genes 3, no. 4: 702-741. https://0-doi-org.brum.beds.ac.uk/10.3390/genes3040702




