Systematic Analysis of the 4-Coumarate:Coenzyme A Ligase (4CL) Related Genes and Expression Profiling during Fruit Development in the Chinese Pear
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Identification and Collection
2.2. RNA Extraction and qRT-PCR Analysis
2.3. Expression Analysis of At4CL/ACS and Os4CL/ACS Genes
3. Results and Discussion
3.1. The Phylogenetic Analysis of Pear 4CL-Related Genes
3.2. Structural Analysis of Pear 4CL-Related Genes
3.3. Collinearity Analyses
3.4. Genomic Distribution and Gene Duplication
3.5. Analysis of Conserved Motifs in Pb4CL and PbACS and Their Promoter Regions
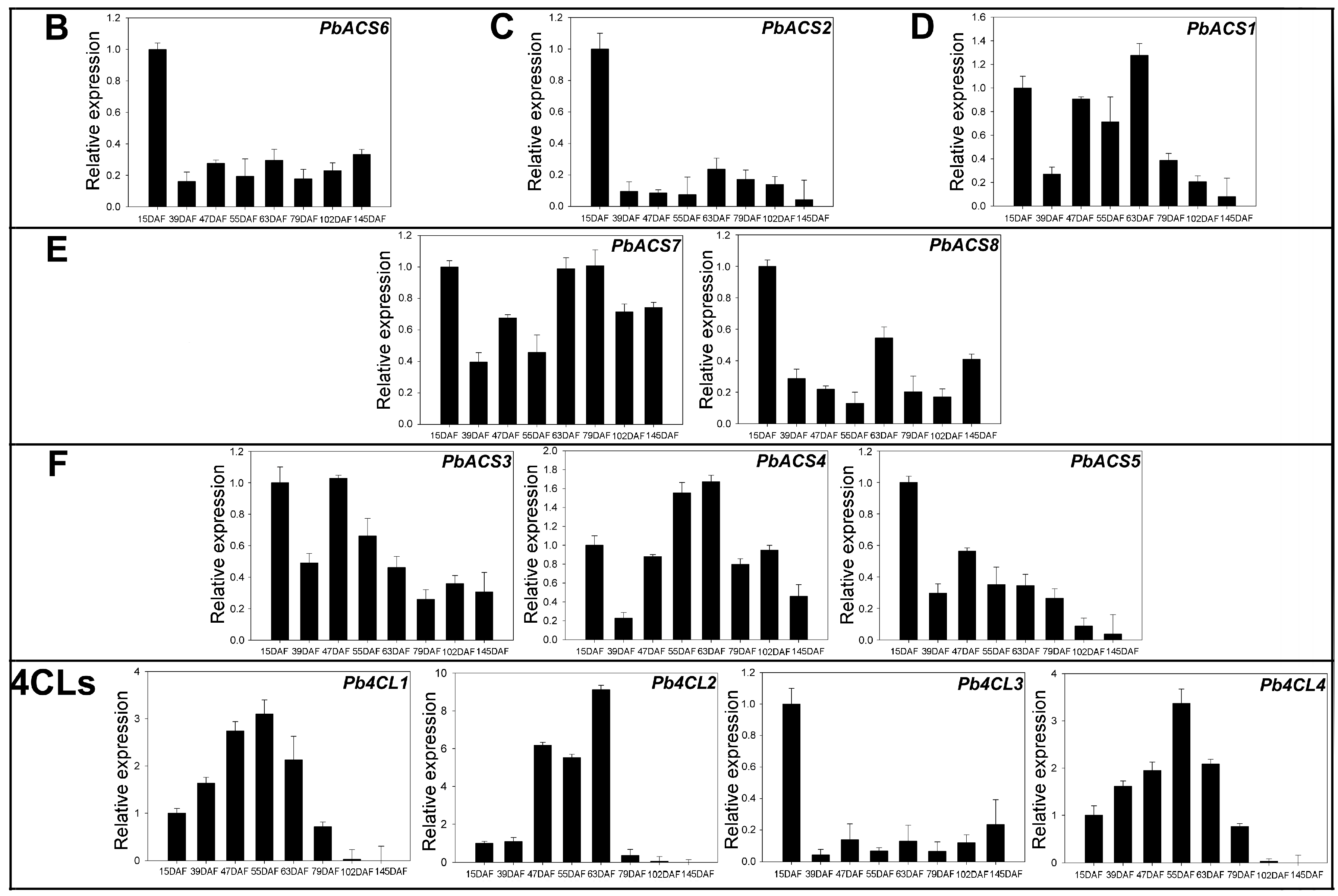
3.6. Expression of 4CLs and ACSs during Ripening in Pear Fruit
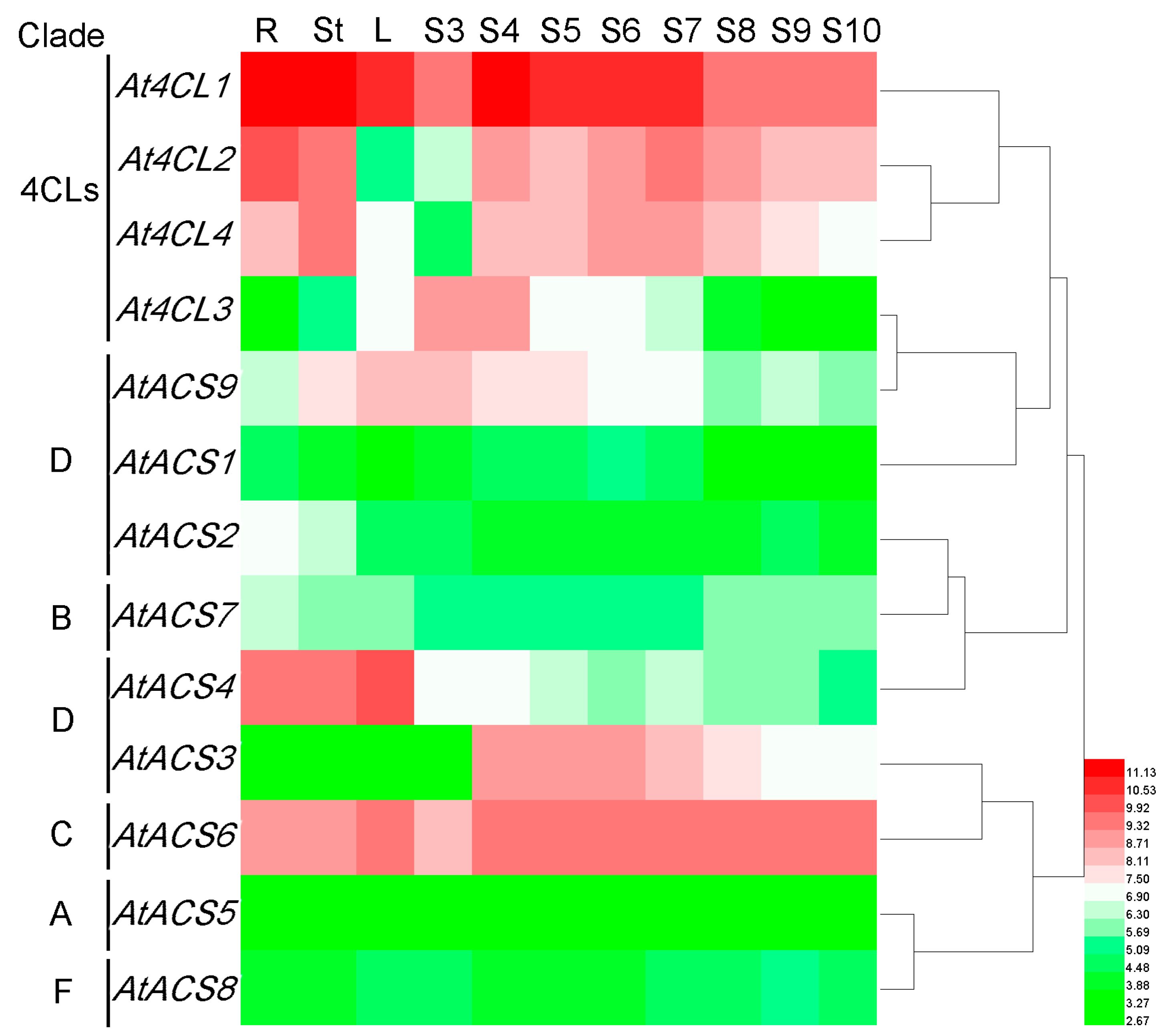
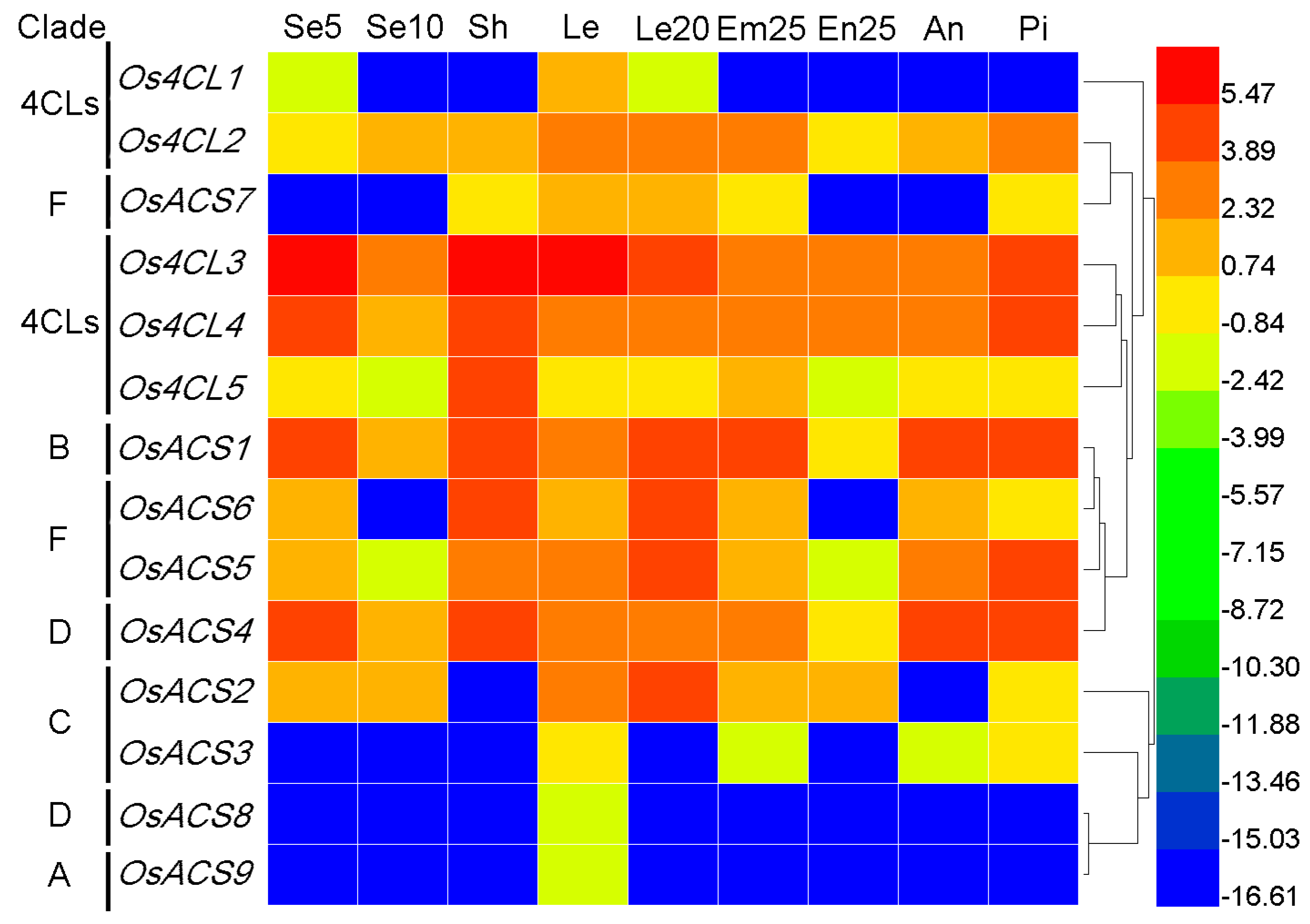
3.7. Expression Profiles of Rice and Arabidopsis 4CL and ACS Genes
3.7.1. Arabidopsis 4CL and ACS Genes
3.7.2. Rice 4CL and ACS Genes
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995, 7, 1085. [Google Scholar] [CrossRef] [PubMed]
- Hamberger, B.; Hahlbrock, K. The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 2004, 101, 2209–2214. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; Harding, S.A.; Tschaplinski, T.J.; Lindroth, R.L.; Yuan, Y. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol. 2006, 172, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Tyler, B. The genome of black cottonwood, Populus trichocarpa. Science 2006, 313, 1596–1604. [Google Scholar]
- Becker-André, M.; Schulze-Lefert, P.; Hahlbrock, K. Structural comparison, modes of expression, and putative cis-acting elements of the two 4-coumarate:CoA ligase genes in potato. J. Biol. Chem. 1991, 266, 8551–8559. [Google Scholar] [PubMed]
- Douglas, C.; Hoffmann, H.; Schulz, W.; Hahlbrock, K. Structure and elicitor or u.v.-light-stimulated expression of two 4-coumarate:CoA ligase genes in parsley. EMBO J. 1987, 6, 1189–1195. [Google Scholar] [PubMed]
- Allina, S.M.; Prihadash, A.; Theilmann, D.A.; Ellis, B.E.; Douglas, C.J. 4-Coumarate:coenzyme A ligase in hybrid poplar. Plant Physiol. 1998, 116, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.J.; Chiang, V.L. Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc. Natl. Acad. Sci. USA 1998, 95, 5407–5412. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Douglas, C.J. Two divergent members of a tobacco 4-coumarate:coenzyme A ligase (4CL) gene family. cDNA structure, gene inheritance and expression, and properties of recombinant proteins. Plant Physiol. 1996, 112, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, A.; Ebel, J. Molecular cloning and expression of 4-coumarate:coenzyme A ligase, an enzyme involved in the resistance response of soybean (Glycine max L.) against pathogen attack. Plant Physiol. 1993, 102, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Harding, S.A.; Leshkevich, J.; Chiang, V.L.; Tsai, C.-J. Differential substrate inhibition couples kinetically distinct 4-coumarate:coenzyme A ligases with spatially distinct metabolic roles in quaking aspen. Plant Physiol. 2002, 128, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Cukovica, D.; Ehlting, J.; Ziffle, J.A.V.; Douglas, C.J. Structure and evolution of 4-coumarate:coenzyme A ligase (4CL) gene families. Biol. Chem. 2001, 382, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Shin, J.J.; Douglas, C.J. Identification of 4-coumarate:coenzyme A ligase (4CL) substrate recognition domains. Plant J. 2001, 27, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Hövel, K.; Witzel, K.; Hamberger, B.; Schomburg, D.; Kombrink, E.; Stuible, H.-P. The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc. Natl. Acad. Sci. USA 2003, 100, 8601–8606. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Mattheus, N.; Aeschliman, D.S.; Li, E.; Hamberger, B.; Cullis, I.F.; Zhuang, J.; Kaneda, M.; Mansfield, S.D.; Samuels, L. Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 2005, 42, 618–640. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.A.; Collins, R.E.; Anterola, A.M.; Cochrane, F.C.; Davin, L.B.; Lewis, N.G. An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry 2003, 64, 1097–1112. [Google Scholar] [CrossRef]
- Shockey, J.M.; Fulda, M.S. Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme a synthetases. Plant Physiol. 2003, 132, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.A.; Bedgar, D.L.; Moinuddin, S.G.; Kim, K.-W.; Cardenas, C.L.; Cochrane, F.C.; Shockey, J.M.; Helms, G.L.; Amakura, Y.; Takahashi, H. Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: Syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 2005, 66, 2072–2091. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Kienow, L.; Schmelzer, E.; Colby, T.; Bartsch, M.; Miersch, O.; Wasternack, C.; Kombrink, E.; Stuible, H.-P. A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J. Biol. Chem. 2005, 280, 13962–13972. [Google Scholar] [CrossRef] [PubMed]
- Kienow, L.; Schneider, K.; Bartsch, M.; Stuible, H.-P.; Weng, H.; Miersch, O.; Wasternack, C.; Kombrink, E. Jasmonates meet fatty acids: Functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J. Exp. Bot. 2008, 59, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Schilmiller, A.L.; Liu, G.; Lee, G.I.; Jayanty, S.; Sageman, C.; Vrebalov, J.; Giovannoni, J.J.; Yagi, K.; Kobayashi, Y. Role of β-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell. 2005, 17, 971–986. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J.; Chung, H.S.; Kobayashi, Y.; Howe, G.A. Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 2006, 281, 33511–33520. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, G.; Nie, J.; Lin, Y.; Nie, F.; Zhang, J.; Xu, Y. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 2010, 125, 374–379. [Google Scholar] [CrossRef]
- Jin, Q.; Yan, C.; Qiu, J.; Zhang, N.; Lin, Y.; Cai, Y. Structural characterization and deposition of stone cell lignin in Dangshan Su pear. Sci. Hortic. 2013, 155, 123–130. [Google Scholar] [CrossRef]
- Pereira, I.; Fachinello, J.; Antunes, L.; Errea, P.; Messias, R.; Pina, A. Expression of the 4-coumarate:CoA ligase gene family in compatible and incompatible Prunus grafts. Acta Hortic. 2013, 976, 333–338. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, W.; Sun, L.; Zhao, F.; Huang, B.; Yang, W.; Tao, Y.; Wang, J.; Yuan, Z.; Fan, G.; et al. The genome of Prunus mume. Nat. Commun. 2012, 3, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D. The genome of the domesticated apple (Malus × Domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nature Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J. The pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S. DNATOOLS, a Software Package for Sequence Analysis; The Carlsberg Laboratory: Copenhagen, Denmark, 2001. [Google Scholar]
- Letunic, I.; Doerks, T.; Bork, P. Smart 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- De Azevedo Souza, C.; Barbazuk, B.; Ralph, S.G.; Bohlmann, J.; Hamberger, B.; Douglas, C.J. Genome-wide analysis of a land plant-specific acyl:coenzyme A synthetase (ACS) gene family in Arabidopsis, poplar, rice and Physcomitrella. New Phytol. 2008, 179, 987–1003. [Google Scholar]
- Hu, B.; Jin, J.; Guo, Y.A.; Zhang, H.; Luo, J.; Gao, G. Gsds 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wang, X.; Bowers, J.E.; Ming, R.; Alam, M.; Paterson, A.H. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008, 18, 1944–1954. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (place) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhang, R.; Gu, C.; Wu, J.; Wan, H.; Zhang, S.; Zhang, S. Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. Afr. J. Agric. Res. 2012, 7, 3701–3704. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, H.; Kapushesky, M.; Kolesnikov, N.; Rustici, G.; Shojatalab, M.; Abeygunawardena, N.; Berube, H.; Dylag, M.; Emam, I.; Farne, A. Arrayexpress update—From an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009, 37, D868–D872. [Google Scholar] [CrossRef] [PubMed]
- Wise, R.A.; Teeter, J.G.; Jensen, R.L.; England, R.D.; Schwartz, P.F.; Giles, D.R.; Ahrens, R.C.; MacIntyre, N.R.; Riese, R.J.; Crapo, R.O. Standardization of the single-breath diffusing capacity in a multicenter clinical trial. CHEST J. 2007, 132, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.-H.; Dardick, C.; Bartley, L.E.; Cao, P.; Phetsom, J.; Canlas, P.; Seo, Y.-S.; Shultz, M.; Ouyang, S.; Yuan, Q. Refinement of light-responsive transcript lists using rice oligonucleotide arrays: Evaluation of gene-redundancy. PLoS ONE 2008, 3, e3337. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. Hemi: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.E.; Greville, K.; Murphy, E.C.; Hooks, M.A. Characterization of Arabidopsis fluoroacetate-resistant mutants reveals the principal mechanism of acetate activation for entry into the glyoxylate cycle. J. Biol. Chem. 2005, 280, 2780–2787. [Google Scholar] [CrossRef] [PubMed]
- Blenda, A.; Zheng, P.; Yu, J.; Bombarely, A.; Cho, I.; Ru, S. The genome database for rosaceae (gdr): Year 10 update. Nucleic Acids Res. 2014, 42, 1237–1244. [Google Scholar]
- Hamberger, B.; Ellis, M.; Friedmann, M.; de Azevedo Souza, C.; Barbazuk, B.; Douglas, C.J. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: The populus lignin toolbox and conservation and diversification of angiosperm gene families. Botany 2007, 85, 1182–1201. [Google Scholar]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef] [PubMed]
- Endler, A.; Martens, S.; Wellmann, F.; Matern, U. Unusually divergent 4-coumarate:CoA-ligases from Ruta graveolens L. Plant Mol. Biol. 2008, 67, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.-X.; Wang, X.-Q. Evolution of 4-coumarate:coenzyme A ligase (4CL) gene and divergence of Larix (Pinaceae). Mol. Phylogenet. Evol. 2004, 31, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.-H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mao, L.; Wang, H.; Brocker, C.; Yin, X.; Vasiliou, V.; Fei, Z.; Wang, X. Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PLoS ONE 2012, 7, e32153. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.-H.; Zhang, S.-Z.; Wang, R.-K.; Zhang, R.-F.; Hao, Y.-J. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 2013, 8, e69955. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.P.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. MYB transcription factors in chinese pear (Pyrus bretschneideri Rehd.): Genome-wide identification, classification and expression profiling during fruit development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, J.A.; Maere, S.; Van de Peer, Y. Plants with double genomes might have had a better chance to survive the cretaceous–tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Cavalcanti, A.; Chen, F.-C.; Bouman, P.; Li, W.-H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhou, Y.; Jiang, H.; Li, X.; Gan, D.; Peng, X.; Zhu, S.; Cheng, B. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS ONE 2011, 6, e28488. [Google Scholar] [CrossRef] [PubMed]
- Han, G.W.; Bakolitsa, C.; Miller, M.D.; Kumar, A.; Carlton, D.; Najmanovich, R.J.; Abdubek, P.; Astakhova, T.; Axelrod, H.L.; Chen, C. Structures of the first representatives of Pfam family PF06938 (DUF1285) reveal a new fold with repeated structural motifs and possible involvement in signal transduction. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29a gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Logemann, E.; Parniske, M.; Hahlbrock, K. Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc. Natl. Acad. Sci. USA 1995, 92, 5905–5909. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Kimura, S.; Demura, T.; Takeda, J.; Ozeki, Y. Dcmyb1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase gene (DcPAL1) in response to elicitor treatment, UV-B irradiation and the dilution effect. Plant Mol. Biol. 2005, 59, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Zhengbradley, X.; Rung, J.; Parkinson, H.; Brazma, A. Large scale comparison of global gene expression patterns in human and mouse. Genome Biol. 2010, 11, 79–82. [Google Scholar]
- Davidson, R.M.; Gowda, M.; Moghe, G.; Lin, H.; Vaillancourt, B.; Shiu, S.H.; Jiang, N.; Buell, C.R. Comparative transcriptomics of three poaceae species reveals patterns of gene expression evolution. Plant J. 2012, 71, 492–502. [Google Scholar] [CrossRef] [PubMed]
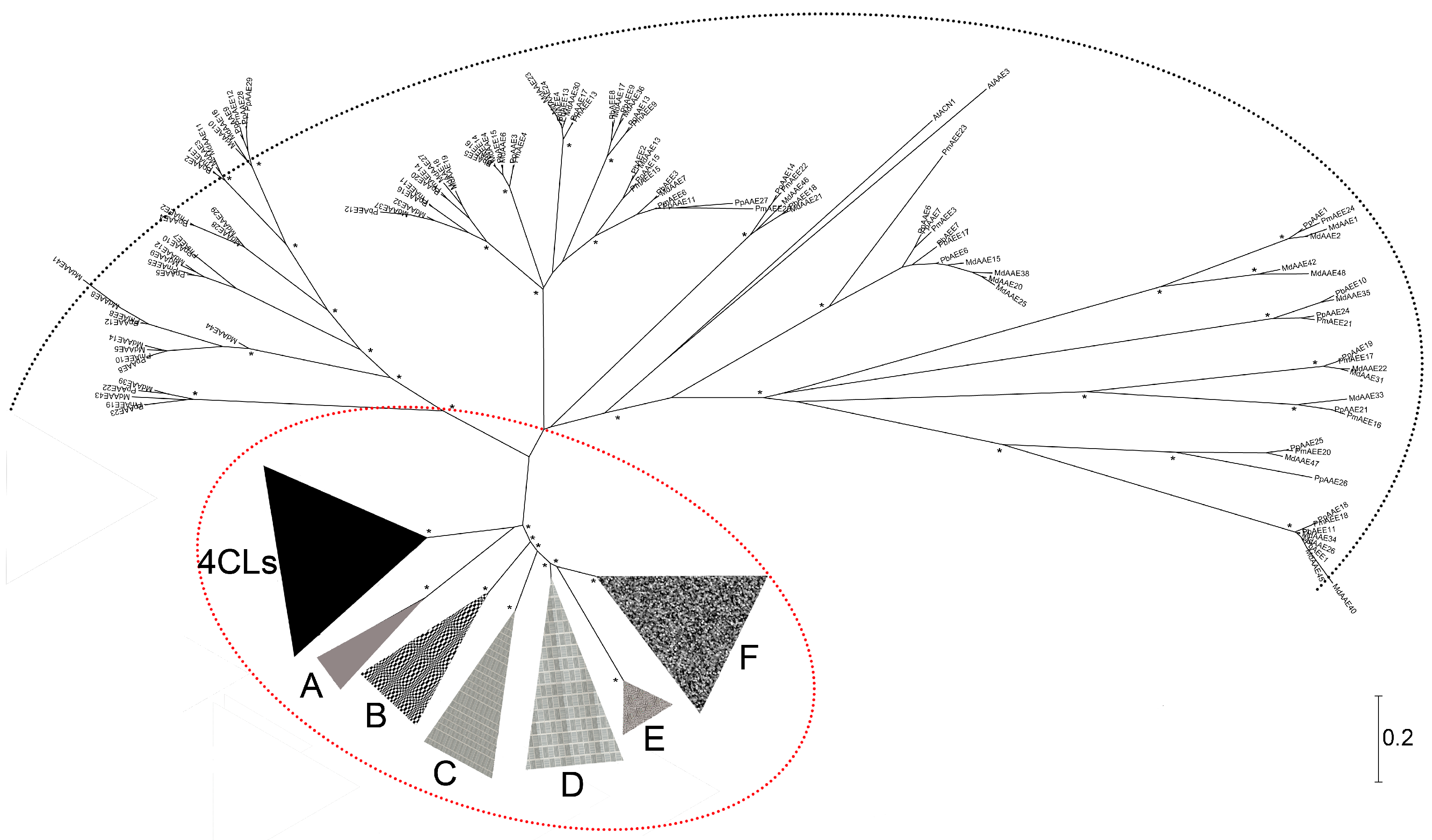
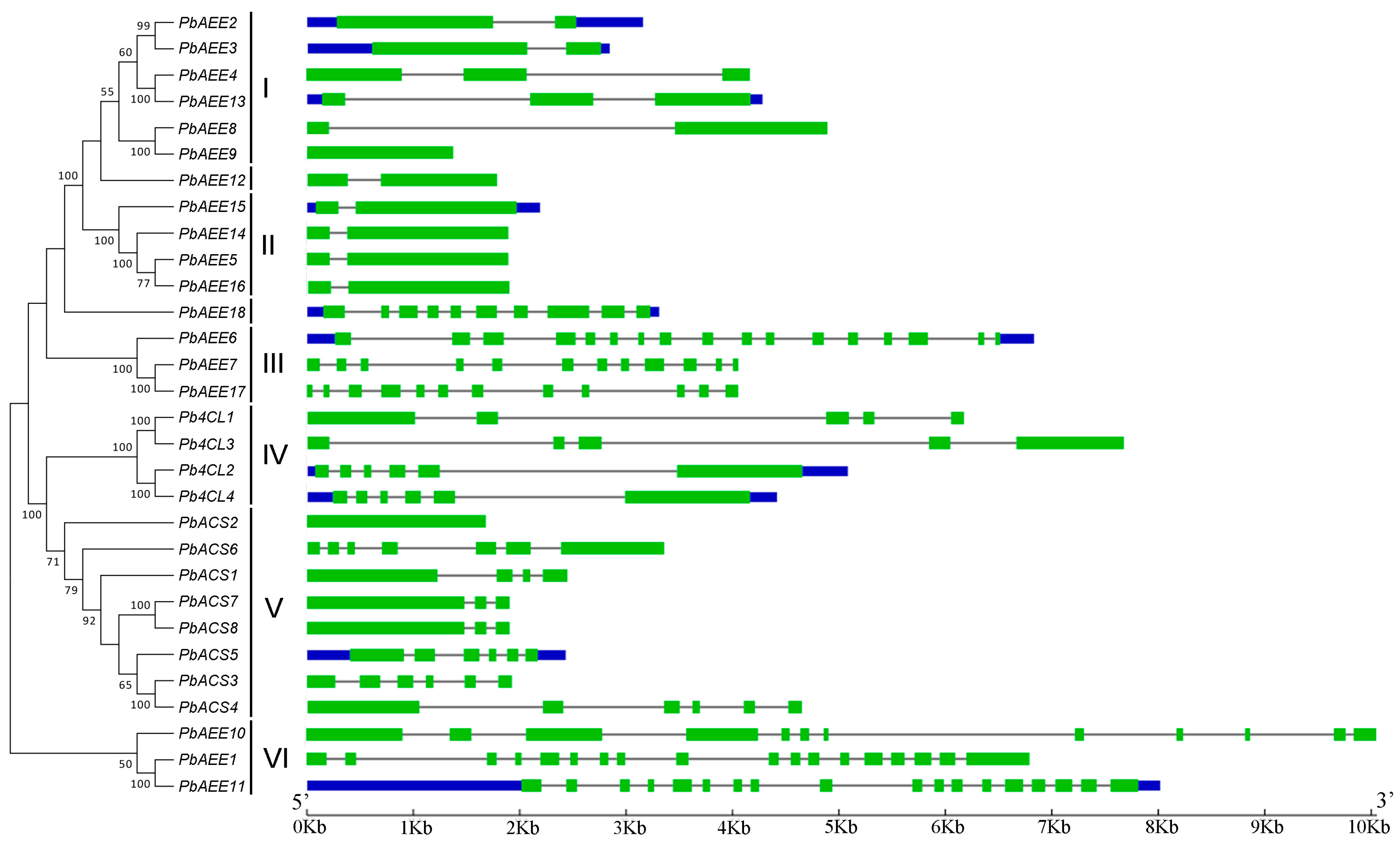
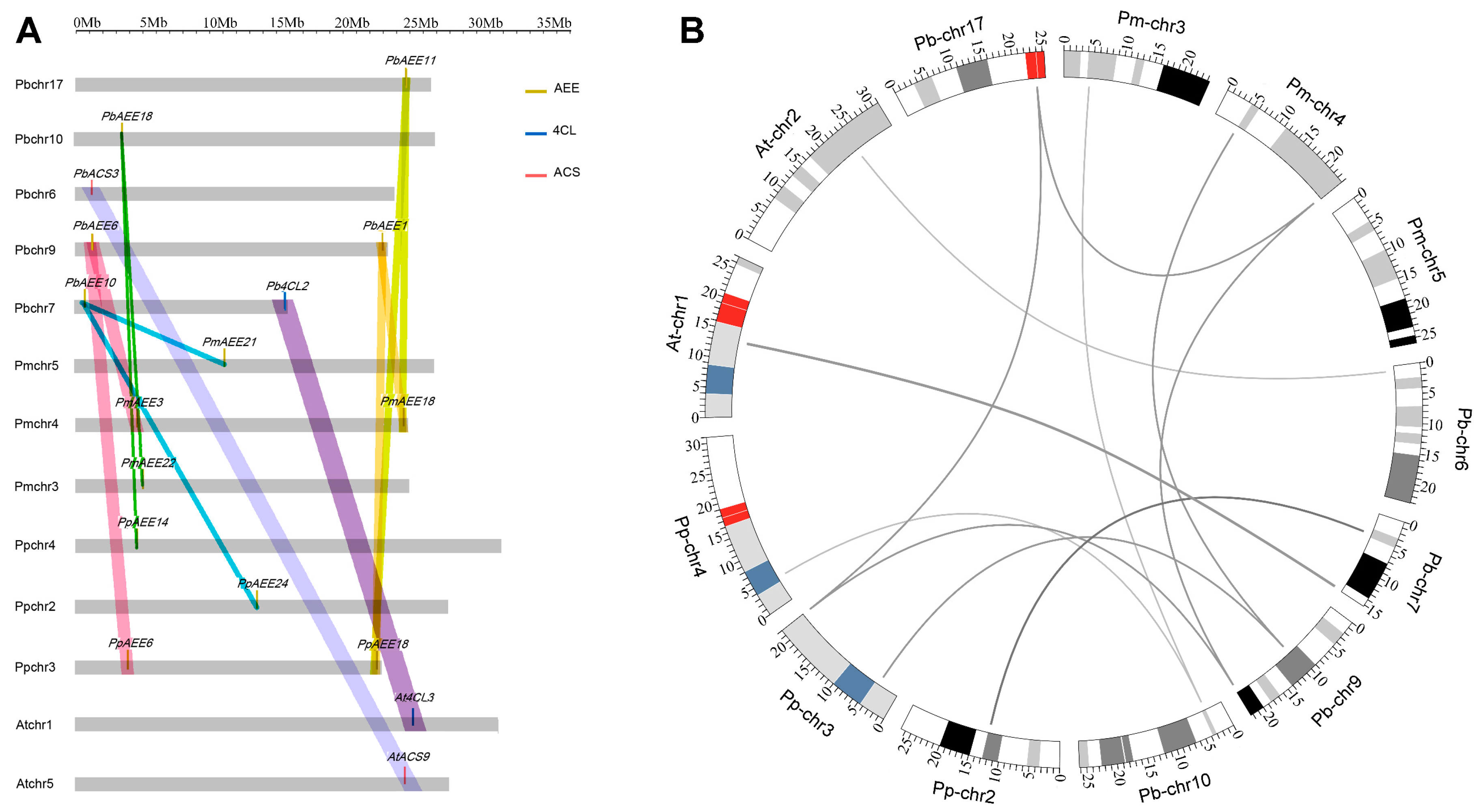

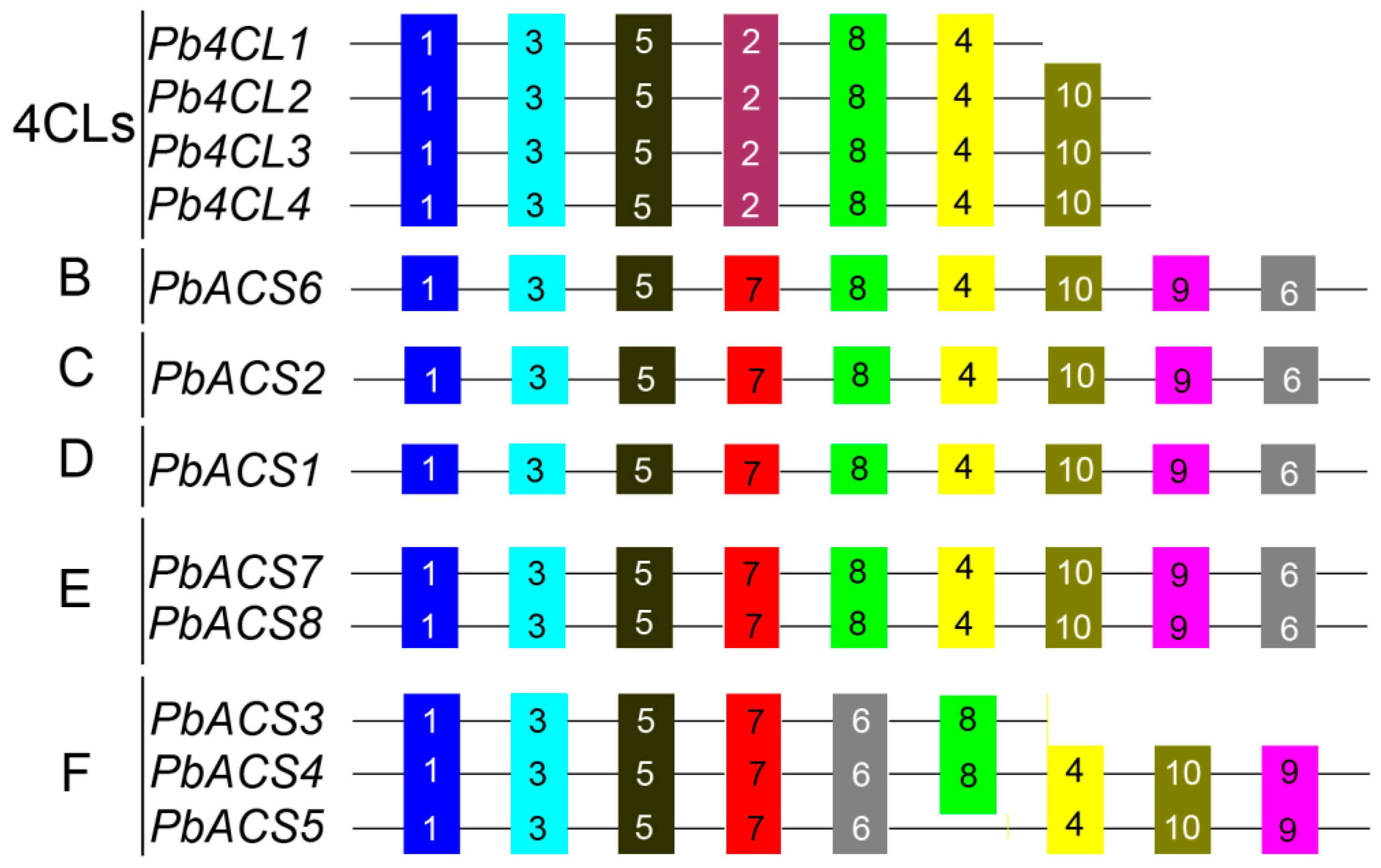
| Name | Clade 1 | Gene Model | Name | Clade 1 | Gene Model |
|---|---|---|---|---|---|
| Pyrus bretschneideri | Prunus mume | ||||
| Pb4CL1 | 4CLs | Pbr024635.1 2 | PmACS5 | D | Pm006110 |
| Pb4CL2 | 4CLs | Pbr039972.1 | PmACS6 | F | Pm024403 |
| Pb4CL3 | 4CLs | Pbr001283.1 | PmACS7 | F | Pm024473 |
| Pb4CL4 | 4CLs | Pbr013445.1 | PmACS8 | A | Pm007022 |
| PbACS1 | D | Pbr005761.1 | PmACS9 | F | Pm024474 |
| PbACS2 | C | Pbr026213.1 | PmACS10 | F | Pm006911 |
| PbACS3 | F | Pbr027219.1 | Prunus persica | ||
| PbACS4 | F | Pbr036926.1 | Pp4CL1 | 4CLs | ppa003747m 5 |
| PbACS5 | F | Pbr036272.1 | Pp4CL2 | 4CLs | ppa003854m |
| PbACS6 | B | Pbr026415.1 | Pp4CL3 | 4CLs | ppa022401m |
| PbACS7 | E | Pbr040326.1 | PpACS1 | B | ppa003893m |
| PbACS8 | E | Pbr041161.1 | PpACS2 | C | ppa017093m |
| Malus × domestica | PpACS3 | C | ppa022562m | ||
| Md4CL1 | 4CLs | MDP0000260512 3 | PpACS4 | D | ppa003506m |
| Md4CL2 | 4CLs | MDP0000295794 | PpACS5 | B | ppa003871m |
| Md4CL3 | 4CLs | MDP0000293578 | PpACS6 | E | ppa022057m |
| Md4CL4 | 4CLs | MDP0000691789 | PpACS7 | F | ppa003658m |
| MdACS1 | A | MDP0000257967 | PpACS8 | F | ppa003674m |
| MdACS2 | D | MDP0000716496 | PpACS9 | A | ppa003797m |
| MdACS3 | E | MDP0000171033 | PpACS10 | F | ppa003742m |
| MdACS4 | D | MDP0000376256 | Oryza sativa | ||
| MdACS5 | E | MDP0000425292 | Os4CL1 | 4CLs | Os08g14760 6 |
| MdACS6 | E | MDP0000206055 | Os4CL2 | 4CLs | Os02g46970 |
| MdACS7 | B | MDP0000277093 | Os4CL3 | 4CLs | Os02g08100 |
| MdACS8 | B | MDP0000267064 | Os4CL4 | 4CLs | Os06g44620 |
| MdACS9 | F | MDP0000179301 | Os4CL5 | 4CLs | Os08g34790 |
| MdACS10 | F | MDP0000289629 | OsACS1 | B | Os03g05780 |
| MdACS11 | A | MDP0000878279 | OsACS2 | C | Os10g42800 |
| MdACS12 | F | MDP0000129547 | OsACS3 | C | Os08g04770 |
| MdACS13 | F | MDP0000262301 | OsACS4 | D | Os03g04000 |
| MdACS14 | F | MDP0000284973 | OsACS5 | F | Os01g67530 |
| MdACS15 | F | MDP0000915947 | OsACS6 | F | Os01g67540 |
| MdACS16 | F | MDP0000206737 | OsACS7 | F | Os07g17970 |
| MdACS17 | F | MDP0000671749 | OsACS8 | D | Os07g44560 |
| MdACS18 | F | MDP0000278716 | OsACS9 | A | Os04g24530 |
| MdACS19 | F | MDP0000303675 | Arabidopsis | ||
| MdACS20 | B | MDP0000300934 | At4CL1 | 4CLs | At1g51680 7 |
| MdACS21 | F | MDP0000282038 | At4CL2 | 4CLs | At3g21240 |
| MdACS22 | F | MDP0000275090 | At4CL3 | 4CLs | At1g65060 |
| MdACS23 | D | MDP0000249003 | At4CL4 | 4CLs | At3g21230 |
| MdACS24 | F | MDP0000723168 | AtACS1 | D | At1g20480 |
| MdACS25 | A | MDP0000249364 | AtACS2 | D | At1g20490 |
| Prunus mume | AtACS3 | D | At1g20500 | ||
| Pm4CL1 | 4CLs | Pm013887 4 | AtACS4 | D | At1g20510 |
| Pm4CL2 | 4CLs | Pm019600 | AtACS5 | A | At1g62940 |
| Pm4CL3 | 4CLs | Pm008736 | AtACS6 | C | At4g05160 |
| PmACS1 | C | Pm021042 | AtACS7 | B | At4g19010 |
| PmACS2 | C | Pm026124 | AtACS8 | F | At5g38120 |
| PmACS3 | B | Pm026358 | AtACS9 | D | At5g63380 |
| PmACS4 | E | Pm017552 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. Systematic Analysis of the 4-Coumarate:Coenzyme A Ligase (4CL) Related Genes and Expression Profiling during Fruit Development in the Chinese Pear. Genes 2016, 7, 89. https://0-doi-org.brum.beds.ac.uk/10.3390/genes7100089
Cao Y, Han Y, Li D, Lin Y, Cai Y. Systematic Analysis of the 4-Coumarate:Coenzyme A Ligase (4CL) Related Genes and Expression Profiling during Fruit Development in the Chinese Pear. Genes. 2016; 7(10):89. https://0-doi-org.brum.beds.ac.uk/10.3390/genes7100089
Chicago/Turabian StyleCao, Yunpeng, Yahui Han, Dahui Li, Yi Lin, and Yongping Cai. 2016. "Systematic Analysis of the 4-Coumarate:Coenzyme A Ligase (4CL) Related Genes and Expression Profiling during Fruit Development in the Chinese Pear" Genes 7, no. 10: 89. https://0-doi-org.brum.beds.ac.uk/10.3390/genes7100089






