Effect of Genetic Diversity in Swine Leukocyte Antigen-DRA Gene on Piglet Diarrhea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Collection and DNA Extraction
2.2. PCR Amplification and Single-Stranded Conformational Polymorphism (SSCP) Analysis
2.3. Cloning and Sequencing
2.4. Sequence and Statistical Analysis
3. Results
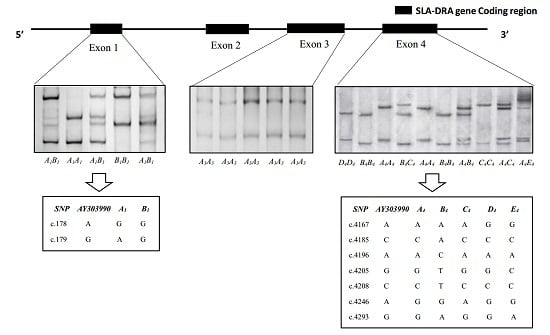
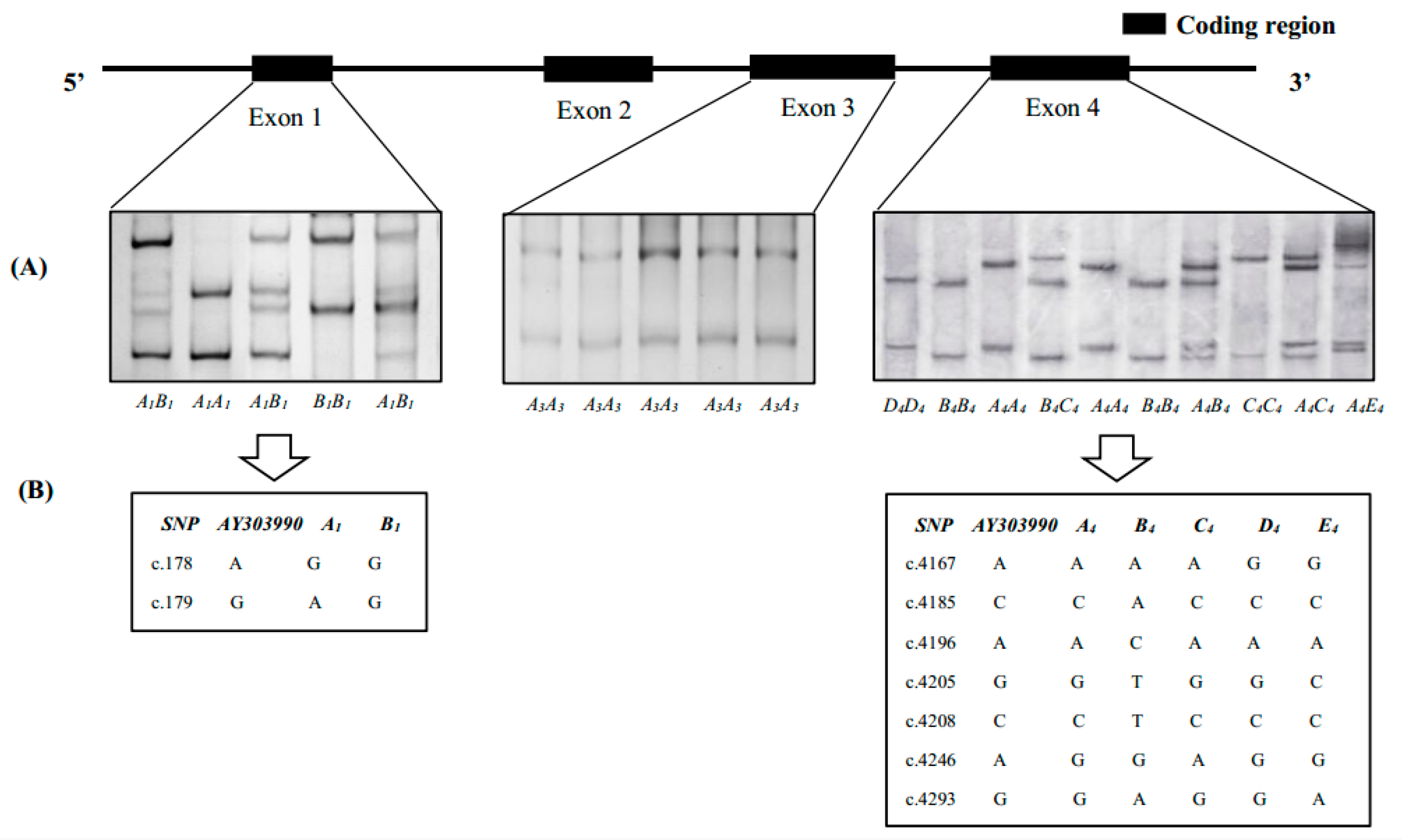
3.1. SSCP Analysis of SLA-DRA Gene Exons
3.2. Gene Variations and Population Genetic Parameters of SLA-DRA Gene
3.3. Amino Acid Alignments of the DRA Gene
3.4. Effects of SLA-DRA Gene Genotypes on Piglet Diarrhea
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lunney, J.K.; Ho, C.S.; Wysocki, M.; Smith, D.M. Molecular genetics of the swine major histocompatibility complex, the SLA complex. Dev. Comp. Immunol. 2009, 33, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.P.; Rohrer, G.A.; Alexander, L.J.; Troyer, D.L.; Kirby-Dobbels, K.R.; Janzen, M.A.; Cornwell, D.L.; Louis, C.F.; Schook, L.B.; Beattie, C.W. Directed integration of the physical and genetic linkage maps of the swine chromosome 7 reveals that SLA spans the centromere. Genom. Res. 1995, 5, 259–271. [Google Scholar] [CrossRef]
- Piriou-Guzylack, L.; Salmon, H. Membrane markers of the immune cells in swine: an update. Vet. Res. 2008, 39, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.; Germana, S.; Gustafsson, K.; Pratt, K.; Sachs, D.H.; Leguern, C. Structure and expression of class II alpha genes in miniature swine. J. Immunol. 1992, 149, 841–846. [Google Scholar] [PubMed]
- Zhou, H.; Hickford, J.G.H.; Fang, Q.; Byun, S.O. Identification of allelic variation at the bovine DRA locus by polymerase chain reaction-single strand conformational polymorphism. J. Dairy Sci. 2007, 90, 1943–1946. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zheng, H.; Xi, D.; Zhang, X.; Du, M.; Pu, L.; Lin, M.; Yang, Y. Molecular characteristics of the MHC-DRA genes from yak (Bos grunniens) and Chinese yakow (Bos grunniens × Bos taurus). Int. J. Immunogenet. 2014, 41, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Zhang, H.L.; Xiang, R.R.; Zhang, Z.W.; Ling, F.; Zhuo, M.; Du, H.L.; Wang, X.N. Identification of Mamu-DPA1, Mamu-DQA1, and Mamu-DRA alleles in a cohort of Chinese rhesus macaques. Immunogenetics 2013, 65, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Arbanasić, H.; Galov, A.; Ambriović-Ristov, A.; Grizelj, J.; Arsenos, G.; Marković, B.; Dovenski, T.; Vince, S.; Curik, I. Extensive polymorphism of the major histocompatibility complex DRA gene in Balkan donkeys: Perspectives on selection and genealogy. Anim. Genet. 2013, 44, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Kamath, P.L.; Getz, W.M. Adaptive molecular evolution of the Major Histocompatibility Complex genes, DRA and DQA, in the genus Equus. BMC Evol. Biol. 2011, 11, 128. [Google Scholar] [CrossRef] [PubMed]
- Keith, T.; Ballingall, M.S.; Rocchi, D.J.; McKeever, F.W. Trans-Species Polymorphism and Selection in the MHC Class II DRA Genes of Domestic Sheep. PLoS ONE 2010, 5, e11402. [Google Scholar]
- Schaid, D.J.; Spraggs, C.F.; Mcdonnell, S.K.; Parham, L.R.; Cox, C.J.; Ejlertsen, B.; Finkelstein, D.M.; Rappold, E.; Curran, J.; Cardon, L.R.; et al. Prospective validation of HLA- DRB1* 07: 01 allele carriage as a predictive risk factor for lapatinib-induced liver injury. J. Clin. Oncol. 2014, 32, 2296–2302. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Han, G.; Jia, B.; Jiang, S.; Du, Y. MHC-DRB1/DQB1 gene polymorphism and its association with resistance/susceptibility to cystic Echinococcosis in Chinese merino sheep. J. Parasitol. Res. 2014, 2–3, 272601. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Han, L.; Han, J.; Liu, J.; Jiang, Q.; Guo, D.; Qu, L. Establishment of six homozygous MHC-B haplotype populations associated with susceptibility to Marek’s disease in Chinese specific pathogen-free BWEL chickens. Infect. Genet. Evol. 2015, 29, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.L.; Takeshima, S.N.; Ohno, A.; Matsumoto, Y.K.; Isogai, Y.; Kohara, Y.K.; Aida, Y.K. Epitope mapping of CD8+ T cells on bovine leukemia virus Gag, Env and Tax protein in cattle with different bovine MHC DRB3 alleles. Retrovirology 2015, 12. [Google Scholar] [CrossRef]
- Hickford, J.G.H.; Forrest, R.H.; Zhou, H.; Fang, Q.; Frampton, C.M. Association between variation in faecal egg count for a mixed field-challenge of nematode parasites and ovine MHC-DQA2 polymorphism. Vet. Immunol. Immunopath. 2011, 144, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Gelasakis, A.I.; Arsenos, G.; Hickford, J.; Zhou, H.; Psifidi, A.; Valergakis, G.E.; Banos, G. Polymorphism of the MHC-DQA2 gene in the Chios dairy sheep population and its association with footrot. Livest. Sci. 2013, 153, 56–59. [Google Scholar] [CrossRef]
- Liu, L.X.; Zhao, S.G.; Lu, H.N.; Yang, Q.L.; Huang, X.Y.; Gun, S.B. Association between polymorphisms of the swine MHC-DQA gene and diarrhoea in three Chinese native piglets. Int. J. Immunogenet. 2015, 42, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.L.; Kong, J.J.; Wang, D.W.; Zhao, S.G.; Gun, S.B. Swine leukocyte antigen-DQA gene variation and its association with piglet diarrhea in Large White, Landrace and Duroc. Asian Australs. J. Anim. Sci. 2013, 26, 1065–1071. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.L.; Zhao, S.G.; Wang, D.W.; Feng, Y.; Jiang, T.T.; Huang, X.Y.; Gun, S.B. Association between Genetic Polymorphism in the swine leukocyte antigen-DRA gene and piglet diarrhea in three Chinese pig breeds. Asian Australs. J. Anim. Sci. 2014, 27, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.L.; Huang, X.Y.; Yuan, J.H.; Gun, S.B. Polymorphisms in the SLA-DQA and DRA gene exon2 and their association with piglet diarrhea in Chinese Yantai black pig. Philipp. Agric. Sci. 2015, 98, 253–261. [Google Scholar]
- Huang, X.Y.; Yang, Q.L.; Yuan, J.H.; Gun, S.B. Polymorphism and haplotype analyses of swine leukocyte antigen DQA exons 2, 3, 4, and their associations with piglet diarrhea in Chinese native pig. Genet. Mol. Res. 2015, 14, 10461–10472. [Google Scholar] [CrossRef] [PubMed]
- Autista, E.M.; Ferman, G.S.; Gregg, D.; Brum, M.C.S.; Grubman, M.J.; Golde, W.T. Constitutive expression of alpha interferon by skin dendritic cells confers resistance to infection by foot and mouth disease virus. J. Virol. 2005, 79, 4838–4847. [Google Scholar] [CrossRef] [PubMed]
- Tissot, R.G.; Beanie, C.W.; Amoss, M.S.; Williams, J.D.; Schumacher, J. Common swine leucocyte antigen (SLA) haplotypes in NIH and Sinclair miniature swine have similar effects on the expression of an inherited melanoma. Anim. Genet. 1993, 24, 19l–194. [Google Scholar] [CrossRef]
- Molina, R.M.; Cha, S.H.; Chittick, W.; Lawson, S.; Murtaugh, M.P.; Nelson, E.A.; Christopher-Hennings, J.; Yoon, K.J.; Evans, R.; Rowland, R.R.R.; et al. Immune response against porcine reproductive and respiratory syndrome virus during acute and chronic infection. Vet. Immunol. Immunop. 2008, 126, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Wimmers, K.; Schellander, K.; Ponsuksili, S. BF, HP, DQB and DRB are associated with haemolytic complement activity, acute phase protein reaction and antibody response in the pig. Vet. Immunol. Immunop. 2004, 99, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H. The Breed Differences in SLA Haplotypes from the Pigs for Xenotransplantation Researches in Korea. In Proceedings of the Plant and Animal Genome XXII Conference, San Diego, CA, USA, 10–15 January 2014.
- Morris, R.S.; Davies, P.R.; Lawton, D.E. Evolution of diseases in the world’s pig industry. In Proceedings of the 17th International Pig Veterinary Society Congress, Ames, IA, USA, 2–5 June 2002.
- Bao, W.B.; Ye, L.; Pan, Z.Y.; Zhu, J.; Du, Z.D.; Zhu, G.Q.; Huang, X.G.; Wu, S.L. Microarray analysis of differential gene expression in sensitive and resistant pig to Escherichia coli F18. Anim. Genet. 2012, 43, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Verhelst, R.; Schroyen, M.; Buys, N.; Niewold, T. Dietary polyphenols reduce diarrhea in enterotoxigenic Escherichia coli (ETEC) infected post-weaning piglets. Livest. Sci. 2014, 160, 138–140. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Zhao, Q.H.; Zi, C.; Wu, Z.C.; Su, X.M.; Huo, Y.J.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Genetic variation in exon 10 of the BPI gene is associated with Escherichia coli F18 susceptibility in Sutai piglets. Gene 2013, 523, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Reyes, L.M.; Blosser, R.J.; Smith, R.F.; Miner, A.C.; Paris, L.L.; Blankenship, R.L.; Tector, M.F.; Tector, A.J. Characterization of swine leucocyte antigen alleles in a crossbred pig to be used in xenotransplant studies. Tissue Antigens 2014, 84, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, L.E.; Jungersen, G.; Sorensen, M.R.; Ho, C.S.; Vadekær, D.F. Swine Leukocyte Antigen (SLA) class I allele typing of Danish swine herds and identification of commonly occurring haplotypes using sequence specific low and high resolution primers. Vet. Immunol. Immunop. 2014, 162, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Zhou, H.; Hickford, J.G.H. Clonal polymerase chain reaction-single-strand conformational polymorphism analysis: an effective approach for identifying cloned sequences. Anal. Biochem. 2008, 378, 111–112. [Google Scholar] [CrossRef] [PubMed]
- Byun, S.O.; Fang, Q.; Zhou, H.; Hickford, J.G.H. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal. Biochem. 2009, 385, 174–175. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Pharoah, P. Commonly studies single-nucleotide polymorphisms and breast cancer: Results from the breast cancer association consortium. J. Natl. Cancer Inst. 2007, 99. [Google Scholar] [CrossRef]
- Blancher, A.; Aarnink, A.; Yamada, Y.; Tanaka, K.; Yamanaka, H.; Shiina, T. Study of MHC class II region polymorphism in the Filipino cynomolgus macaque population. Immunogenetics 2014, 66, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.X. Molecular Genetic Characteristics of SLA-DQA and DRA Genes and Their Association with Piglet Diarrhea in Pig of China. Ph.D. Thesis, Gansu Agricultural University, Lanzhou, Gansu, China, 2015. [Google Scholar]
- Posada, D.; Crandall, K.A. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol. Evol. 2001, 16, 37–45. [Google Scholar] [CrossRef]
- Ho, C.S.; Lunney, J.K.; Lee, J.H.; Franzo-Romain, M.H.; Martens, G.W.; Rowland, R.R.R.; Smith, D.M. Molecular characterization of swine leucocyte antigen class II genes in outbred pig populations. Anim. Genet. 2010, 41, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Sakaram, D.; Niranjan, S.K.; Kumar, S.; Naskar, S.; Deb, S.M.; Mitra, A.; Sharma, A.; Sharma, D. cDNA characterization and molecular analysis of buffalo MHC class II gene, DRA (Bubu-DRA). J. Appl. Anim. Res. 2010, 37, 73–76. [Google Scholar] [CrossRef]
- Le, M.T.; Choi, H.; Choi, M.K.; Nguyen, D.T.; Kim, J.H.; Seo, H.G.; Cha, S.Y.; Seo, K.; Chun, T.; Schook, L.B.; et al. Comprehensive and high-resolution typing of swine leukocyte antigen DQA from genomic DNA and determination of 25 new SLA class II haplotypes. Tissue Antigens 2012, 80, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Jiang, Q.; Guo, D.; Liu, J.; Han, L.; Qu, L. Characterization of swine leukocyte antigen (SLA) polymorphism by sequence-based and PCR-SSP methods in Chinese Bama miniature pigs. Dev. Comp. Immunol. 2014, 45, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Miyasaka, T.; Takeshima, S.N.; Jimba, M.; Matsumoto, Y.; Kobayashi, N.; Matsuhashi, T.; Sentsui, H.; Aida, Y. Identification of bovine leukocyte antigen class II haplotypes associated with variations in bovine leukemia virus proviral load in Japanese Black cattle. Tissue Antigens 2013, 81, 72–82. [Google Scholar] [CrossRef] [PubMed]

| Region | Location a | Primer F/R(5′→3′) | Amplification Fragment | Annealing Temperature (°C) | SSCP Condition |
|---|---|---|---|---|---|
| Exon 1 | 151–226 | F: CTTTGCTTGTATTGC R: ACCTAACTACCCCTC | 186 bp | 56.8 | 4 °C, 12%, 39:1, 190 V 20 h |
| Exon 3 | 4595–4876 | F: TGCTAAACAGGGAAGGCT R: ACAAAGGAGACTGAGGGATG | 352 bp | 56.8 | 4 °C, 10%, 39:1, 200 V 20 h |
| Exon 4 | 4155–4309 | F: TCCCGTAATACATCGTTC R: TTCCTTTCCTTGGCTCAT | 357 bp | 55.6 | 18 °C, 10%, 29:1, 200 V 18 h |
| Locus | Allele | F (%) A | GenBank Accession Number | AE B | oHet/eHet C | PIC D | χ2 E | GenBank Ident 100% |
|---|---|---|---|---|---|---|---|---|
| Accession Number/Breed | ||||||||
| Exon 1 | A1 | 0.55 | KR023998 | 1.979 | 0.260/0.495 | 0.372 | 65.77 * | JX135565/Landrace |
| B1 | 0.45 | KR023999 | LC002669/Hebao pig FJ905824/Gansu white pig | |||||
| Exon 4 | A4 | 0.41 | KM485553 | 3.087 | 0.439/0.677 | 0.612 | 511.97 * | KP324812/Juema pig FJ905836/Hezuo pig |
| B4 | 0.34 | KM485554 | Not found | |||||
| C4 | 0.20 | KM487705 | KP324810/Juema pig AY247781/Hebao pig | |||||
| D4 | 0.01 | KM487706 | KP324814/Juema pig AY191779/Banna minipig | |||||
| E4 | 0.04 | KM487707 | EU432071/Meishan pig EU722916/CMS minipig DQ883222/Korean native pig |
| Exon 1 | Exon 4 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | SN A | Case | Control | OR B | LSM ± SE C | p | Genotype | SN | Case | Control | OR | LSM ± SE | p |
| A1A1 | 123 | 60 | 63 | 1.140 | 1.00 ± 0.11 | 0.679 | A4A4 | 66 | 26 | 40 | 0.674 | 0.81 ± 0.14 | 0.302 |
| B1B1 | 92 | 44 | 48 | 0.957 | 1.08 ± 0.12 | 0.424 | B4B4 | 80 | 40 | 40 | 0.623 | 0.98 ± 0.13 | 0.462 |
| A1B1 | 75 | 32 | 43 | 0.794 | 0.89 ± 0.14 | 0.348 | A4B4 | 12 | 4 | 8 | 0.553 | 0.80 ± 0.33 | 0.114 |
| C4C4 | 12 | 8 | 4 | 2.344 | 1.80 ± 0.33 | 0.001 | |||||||
| A4C4 | 72 | 32 | 40 | 0.877 | 0.85 ± 0.14 | 0.933 | |||||||
| B4C4 | 22 | 8 | 14 | 0.625 | 0.77 ± 0.25 | 0.715 | |||||||
| A4E4 | 22 | 14 | 8 | 2.094 | 1.66 ± 0.25 | 0.002 | |||||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Yang, Q.; Yuan, J.; Liu, L.; Sun, W.; Jiang, Y.; Zhao, S.; Zhang, S.; Huang, W.; Gun, S. Effect of Genetic Diversity in Swine Leukocyte Antigen-DRA Gene on Piglet Diarrhea. Genes 2016, 7, 36. https://0-doi-org.brum.beds.ac.uk/10.3390/genes7070036
Huang X, Yang Q, Yuan J, Liu L, Sun W, Jiang Y, Zhao S, Zhang S, Huang W, Gun S. Effect of Genetic Diversity in Swine Leukocyte Antigen-DRA Gene on Piglet Diarrhea. Genes. 2016; 7(7):36. https://0-doi-org.brum.beds.ac.uk/10.3390/genes7070036
Chicago/Turabian StyleHuang, Xiaoyu, Qiaoli Yang, Junhu Yuan, Lixia Liu, Wenyang Sun, Yingdi Jiang, Shengguo Zhao, Shengwei Zhang, Wangzhou Huang, and Shuangbao Gun. 2016. "Effect of Genetic Diversity in Swine Leukocyte Antigen-DRA Gene on Piglet Diarrhea" Genes 7, no. 7: 36. https://0-doi-org.brum.beds.ac.uk/10.3390/genes7070036