1. Introduction
Genome stability through multiple cell divisions needs faithful segregation of sister chromatids to the daughter cells. To accomplish this, any linkage that may remain between sisters must have been removed by the time cells enter anaphase. Sister chromatid linkages can be proteinaceous (e.g., cohesion), topological (e.g., catenations) or DNA-DNA mediated (i.e., presence of underreplicated DNA and joint molecules). Joint molecules (JMs) are transient intermediates of the homologous recombination (HR) pathway. In cells undergoing mitotic division, HR works as a reliable error-free DNA repair mechanism against DNA double strand breaks (DSB), stalled replication forks (SRFs), interstrand crosslinks and single stranded DNA (ssDNA) gaps. If sister chromatid linkages are not removed by anaphase, they give rise to anaphase bridges (ABs). In this scenario, the mitotic division may end up in sister chromatid nondisjunction with or without associated DBSs [
1]. DSBs at this cell cycle stage are extremely dangerous for the progeny since both ends of the broken DNA lay in different daughter cells and, therefore, there is no way to restore the original DNA molecule by any means.
Joint molecules arise upon synapsis of one DNA strand with the complementary strand of the sister chromatid and can be the consequence of the invasion of one end coming from a DSB, the switch of the leading strand DNA template to bypass a SRF, or else, the switch of the template to fill up a ssDNA gap in one sister chromatid [
2,
3]. Common to all these variants, the first synaptic event is the formation of a JM termed displacement loop (D-loop). To accomplish synapsis, acceptor DNA must first be single-stranded and coated with the replication protein A (RPA) complex. Then, RPA is displaced by Rad51 to form the ssDNA-Rad51 filament, which starts the search for homology along the donor sister chromatid. Once satisfactory homology is found, the D-loop is shaped by the displacement of the non-complementary donor strand, which then becomes coated by RPA [
4,
5]. Aside from Rad51, there are other key players during these early HR steps. Thus, important catalysts for DNA synapsis in yeast are Rad52 and other members of its epistasis group such as Rad54. Antagonists of the HR synapsis also exist; for instance, the helicase Srs2 is capable of disassembling the Rad51 filament. Once a stable D-loop has been formed, postsynaptic steps complete HR. In all cases, DNA synthesis will be required. In the synthesis dependent strand annealing (SDSA) pathway, the D-loop remains as the priming substrate for copying the information from the donor sister chromatid and then it is dismantled. Alternatively, the D-loop can be further processed into one or two cross-shaped JM termed Holliday junction (HJ). D-loops and HJs can be resolved by endonucleolytic enzymes or, just in the case of D-loops, by the sole action of DNA helicases [
6,
7,
8]. There is, though, a high degree of specificity for the resolution of each structure. Thus, D-loops are disassembled by the helicase Mph1 (Fanconi anaemia complementation group M, FANCM, in humans), whereas HJs can be resolved by specialised structure selective endonucleases (SSEs), and double HJs (dHJ) by a dissolution pathway which involves the helicase-topoisomerase complex Sgs1-Top3-Rmi1 (STR). This complex is homologous to the human Bloom’s syndrome BLM–TopoIIIa–RMI1–RMI2 complex. In yeast, there are three SSEs able to resolve HJs: the heterodimer complexes Mus81-Mms4 (MUS81-EME1 in humans) and Slx1-Slx4 (SLX1-SLX4/FANCP in humans), and the resolvase Yen1 (GEN1 in humans).
In recent years, a bunch of papers in different organisms have shown that mutants for some of these postsynaptic players form ABs [
9,
10,
11]. Interestingly, proteins such as BLM and FANCM also localised to a new type of DAPI-invisible bridges termed ultrafine anaphase bridges (UFBs) [
12,
13]. All these bridges happen to be despite HR is tightly coupled to cell cycle checkpoints that prevent anaphase onset upon DNA damage. For instance, the intra-S phase and G2 DNA damage checkpoints can be triggered by conditions that lead to SRFs (e.g., DNA alkylation by methyl methanesulfonate (MMS)). DSBs can also trigger the G2 DNA damage checkpoint. At present, all evidences argue that the RPA-coated ssDNA, which is present in the early processing steps of DSBs by HR and that can also be formed at SRFs, is the universal signal for checkpoint activation and maintenance [
14,
15]. Nevertheless, since early steps of the HR displace RPA coated ssDNA with Rad51, and then this filament forms JMs upon synapsis, it might be conceptually possible that these checkpoints have been switched off before many JMs are fully resolved. This situation can be exacerbated if cells are challenged to resolve JMs at an increased rate; e.g., during replication stress. Indeed, we previously showed that the double mutant
mms4Δ
yen1Δ, deficient in the endonucleolytic resolution of JMs, give rise to DAPI-stained ABs [
9]. We further showed that the chromosome XII right arm (cXIIr), which carries the hyperrecombinogenic ribosomal DNA (rDNA) array, was a hot spot for sister chromatid nondisjunction and that this could be rescued by deleting
RAD52, demonstrating that the source of nondisjunction were HR-driven JMs. Both DAPI-stained ABs and cXIIr nondisjunction occurred apparently blind to cell cycle checkpoints, even under exogenously induced replicative stress. In the present work, we have quantified the formation of DAPI-stained bridges and cXIIr nondisjunction in mutants for several helicases with roles in HR. We have specifically focused on FANCM-like Mph1 and its genetic interactions with the other helicases. We have found that abrogation of Mph1 does not give rise to aberrant anaphase phenotypes but blocks cells at G2. Nevertheless, Mph1 contributes to diminish post-G2 aberrations seen when the activity of Rad5 and Rad54, two helicase members of the SWI/SNF family of DNA translocases, have been impaired.
3. Results
In previous works, we showed that sister chromatid nondisjunction in a single cell cycle can be precisely detected by fluorescent microscopy in
Saccharomyces cerevisiae, provided that DAPI-stained ABs are checked together with the segregation status of telomeric regions for the chromosome XII right arm (Tel-cXIIr) [
9,
16,
19]. We had reasoned that Tel-cXIIr segregation status was a good indicator for the overall sister chromatid disjunction since cXIIr appears to be the most segregation-challenging chromosome arm in yeast [
19]. This is so, not only because cXIIr is the longest chromosome arm (
Figure 1A), but also because it harbours the rDNA array. The rDNA is prone to sister chromatid linkages since it carries natural SRFs, is enriched in spontaneous JMs and is a hot-spot for catenations due to its high levels of transcription [
1]. In order to accurately measure Tel-cXIIr segregation, and also stabilise DAPI-stained ABs, we took advantage of the
cdc15-2 thermosensitive allele, which blocks the cells in late mitosis without interfering with chromosome segregation [
9,
16].
When the G1-to-telophase cell cycle is combined with increasing concentrations of a drug in a dose-response curve, further information can be obtained about the capability of the drug to elicit cell cycle blocks at G1 (unbudded cells) and G2 (mononucleated dumbbells) [
20]. By comparing G2 blocks with DAPI-stained ABs and cXIIr nondisjunction in those cells able to pass G2, it is possible to have an overall picture of whether a drug, a mutant, or the combination of drugs and mutants result in anaphase abnormalities unnoticed by the G2 cell cycle checkpoints (
Figure 1B). This rationale is the one we previously used to conclude that the double knockout for the SSEs Mms4-Mus81 and Yen1 did not change the cell cycle profile to the replication stress drug MMS, yet it greatly increased the frequency of ABs and cXIIr nondisjunction [
9]. This is also the rationale we have followed here to analyse knockout mutants for several DNA helicases involved in HR, or with suspected roles in dealing with replicative stresses (i.e., Mph1, Rad54, Sgs1, Srs2 and Rad5).
3.1. Replication Stress Does Not Cause Anaphase Aberrations in Cells Deficient for the Fanconi Anaemia Group M-Like DNA Helicase Mph1
We started this work by comparing the
cdc15-2 mph1Δ mutant to our reference
cdc15-2 strain. The
mph1Δ was included in the original screen where SSEs mutants
mms4Δ and
mms4Δ
yen1Δ were studied [
9]. Unlike the SSEs mutants, the G2 arrest profile in
mph1Δ was different to the reference strain (
Figure 2A, upper charts); whereas MMS addition gave a single peak of G2 arrest at 0.01% v/v in the reference
cdc15-2 strain and the SSEs mutants, the G2 arrest was still prevalent in
cdc15-2 mph1Δ at MMS concentrations 3-fold lower (
Figure 2B). This fact made it impossible to compare anaphase figures at 0.004% v/v, as we had done before for the SSEs mutants. Such “borderline” concentration of MMS had been chosen because it was the highest that allowed most cells to enter anaphase from a cytological point of view; that is, equal or greater than 50% of cells were either binucleated dumbbells or with a stretched DAPI bridge across the bud neck. However, the dose-response MMS curve let us set the borderline concentration for
cdc15-2 mph1Δ at 0.001% (
Table 1), and still compared the anaphase figures between borderline concentrations. Strikingly, neither DAPI-stained bridges (<10% of post-G2 cells) nor cXIIr missegregation (<20%) was higher than the corresponding control without MMS (
Figure 2A, lower charts; and 2C). This anaphase profile was not different to that of the reference
cdc15-2 strain, yet significantly different to the SSEs mutants; particularly the
mms4Δ
yen1Δ double mutant at the borderline concentration (~80% of post-G2 cells missegregated cXIIr and ~20% had DAPI bridges).
In addition to the G1-to-telophase cell cycle profile, we compared the long-term sensitivity to MMS of these mutants through spot assays. We observed that
cdc15-2 mph1Δ was only slightly more sensitive than the reference
cdc15-2 strain (
Figure 2D). By contrast,
cdc15-2 mms4Δ
yen1Δ was rather hypersensitive and
cdc15-2 mms4Δ was still more sensitive than
cdc15-2 mph1Δ. The only mild increase in MMS sensitivity of
mph1Δ is likely a consequence of the compensatory effect of bypassing SRFs by the mutagenic translesion synthesis (TLS) pathway, as it has been shown before in this mutant [
21,
22].
3.2. The G2 and Post-G2 Profiles of mph1Δ are Hypostatic to rad51Δ but Additive to rad54Δ
The fact that
cdc15-2 mph1Δ brought about a broader G2 peak suggests that, under replicative stress, a stronger G2 checkpoint is triggered in this mutant; i.e., more RPA-ssDNA could be present as a result of replication fork stalling by MMS. The steady-state levels of RPA-ssDNA are negatively controlled by the efficiency of the transition between the RPA-ssDNA and the Rad51-DNA filaments [
23]. We therefore compared the
cdc15-2 mph1Δ profiles to those seen in the presynaptic
cdc15-2 rad51Δ mutant. We found that the G2 profile in the
cdc15-2 rad51Δ MMS dose-response curve was even broader than that of the
cdc15-2 mph1Δ strain (
Figure 3A, leftmost upper chart;
Table 1). In addition,
rad51Δ, but not
mph1Δ, yielded DAPI bridges in up to 20% of post-G2 cells at the borderline MMS concentration (
Figure 3A, leftmost lower chart, and
Figure 3B). Of note, the morphology of the DAPI-stained bridge in
rad51Δ tended to show two highly interconnected nuclear masses across the bud neck (
Figure 3C), as opposed to the elongated DAPI bridge seen in the
cdc15-2 mms4Δ
yen1Δ mutant. This happened in at least 75% of the observed
cdc15-2 rad51Δ DAPI-bridges. This bow-tie morphology suggests a highly tight anaphase DAPI-stained bridge, as the one observed in
top2-4 cdc15-2 [
24]. Furthermore, cXIIr missegregation alone (i.e., without a concomitant visible DAPI bridge) was rare in this mutant (<10% of post-G2 cells). Similar cell biology phenotypes were seen in
cdc15-2 rad52Δ (data not shown). Finally, the long-term MMS sensitivity showed remarkable differences between
cdc15-2 mph1Δ and
cdc15-2 rad51Δ; with the latter being strongly hypersensitive to MMS (
Figure 3D).
We next included the
cdc15-2 rad54Δ mutant in this analysis. Rad54 is an ATP DNA translocase and an important catalyst in the disassembly of the Rad51 filament once the DNA synapsis with the donor template takes places, hence facilitating the creation of a functional D-loop [
25]. Dose-response MMS profiles for
cdc15-2 rad51Δ and
cdc15-2 rad54Δ were entirely equivalent (
Figure 3A,B). Just regarding the G2 profile, this result suggests that during the replication stress the G2 checkpoint response is highly active even after disassembling the RPA-ssDNA filament in favour of the Rad51-ssDNA filament.
Finally, we compared the double
mph1Δ
rad51Δ and
mph1Δ
rad54Δ mutants with the corresponding single mutants. The segregation defects (bow-tie DAPI bridges), G2 block profiles and long-term hypersensitivity to MMS of
cdc15-2 mph1Δ
rad51Δ were equivalent to those of
cdc15-2 rad51Δ (
Figure 3A,B,D). This equivalence establishes an epistatic relationship that positions Mph1 downstream Rad51 within the same pathway, as well as suggesting that Mph1 would have a compensatory activity that makes the
mph1Δ mutant milder than presynaptic HR mutants to replication stress. The long-term hypersensitivity and G2 profiles were also similar between
cdc15-2 mph1Δ
rad54Δ and
cdc15-2 rad54Δ, but an additive post-G2 effect was observed at 0.0004% MMS (
Figure 3B). This non-epistatic relationship suggests that Mph1 backs up Rad54 in dealing with replication stress intermediates that may compromise mitotic division.
3.3. Abrogation of the Functionally Related Helicase Srs2 Enhances the mph1Δ G2 Block without Causing More Aberrant Post-G2 Figures
Mph1 is a 3′-to-5′ helicase that belongs to the SF2 superfamily. Srs2 is another SF2 3′-to-5′ helicase which has been clearly implicated in HR and could theoretically complement Mph1. Indeed, Srs2 is able to biochemically dismantle D-loops and has a strong negative genetic interaction with Mph1 [
26,
27]. Consequently, we decided to study the G2 and post-G2 profiles of
cdc15-2 mutant derivatives for
SRS2, compare it to
cdc15-2 mph1Δ, and derive their epistatic relationships through the analysis of the
cdc15-2 mph1Δ
srs2Δ double mutant (
Figure 4). We found that the cell cycle profile to increasing MMS concentrations of
cdc15-2 srs2Δ was reminiscent of
cdc15-2; i.e., a G2 peak at 0.01% MMS, although a slight increase in G2 blocks at lower MMS concentrations were also observed (
Figure 4A). However, The G2 profile of the
mph1Δ
srs2Δ double mutant was synergistic, with a borderline at 0.0004% v/v, 3-fold lower than
mph1Δ. By contrast, the post-G2 segregation figures of
mph1Δ
srs2Δ and
srs2Δ were equivalent (
Figure 4A,B), despite
srs2Δ having an increase in cXIIr missegregation relative to
mph1Δ (~30% vs. ~15%). The long-term hypersensitivity to MMS showed a clear and synergistic negative interaction between
mph1Δ and
srs2Δ (
Figure 4C), confirming previous reports [
22,
27,
28].
3.4. The G2 and Post-G2 Profiles of mph1Δ Are Neutral to Those Observed in sgs1Δ
A third SF2 3′-to-5′ helicase implicated in HR is Sgs1, the yeast ortholog of human BLM. Sgs1 works within the Sgs1-Top3-Rmi1 (STR) helicase-topoisomerase complex as a dHJ dissolvasome. As such, it has been implicated in the removal of late JMs, after the maturation of the D-loop into HJs [
29]. However, BLM has also been shown to dissolve D-loops in a mechanism that might complement FANCM/Mph1 activity [
30,
31]. We found that the cell cycle profile to increasing MMS concentrations of
cdc15-2 sgs1Δ was like
cdc15-2; i.e., a single G2 peak at 0.01% MMS (
Figure 5A). The G2 profile of the
mph1Δ
sgs1Δ double mutant was more similar to
mph1Δ than was to
sgs1Δ, whereas the post-G2 segregation figure was more similar to
sgs1Δ at the corresponding MMS borderline concentrations (
Figure 5A,B). Noteworthy,
sgs1Δ showed a clear dose-response profile of anaphase abnormalities, rather reminiscent of that observed for the SSE
mms4Δ
yen1Δ double mutant, but with just half of their values. These cell cycle profiles likely position Sgs1 at a stage wherein timely removal of late JMs is a must to prevent mitotic catastrophe. In addition, they recapitulate our previous findings with SSE mutants, indicating that JMs that serve as substrates for both Sgs1 and SSEs are not sense by the G2 checkpoints [
9]. The facts that (i) there is no extensive G2 peak in
sgs1Δ and (ii) the G2 peak profile of
mph1Δ
sgs1Δ is epistatic to
mph1Δ, situate Mph1 upstream Sgs1 in a linear pathway devoted to bypassing SRFs. In addition, the facts that (i) there is post-G2 segregation defects in
sgs1Δ, not seen in
mph1Δ, and (ii) the post-G2 profile of
mph1Δ
sgs1Δ is epistatic to
sgs1Δ, suggests that Mph1 has no role in removing late JMs, contrary to our initial expectations. Finally, although our epistatic analysis for short-term phenotypes strongly suggests that Sgs1 and Mph1 do not co-work on the same substrates, we observed an additive effect on long-term MMS sensitivity (
Figure 5C). These results suggest that additive fitness defects in the double mutant might arise from successive cell divisions under suboptimal conditions to confront replication stress; e.g., the consequences of the higher cXIIr missegregation in
sgs1Δ could be ameliorated by Mph1 in the following cell cycles, etc.
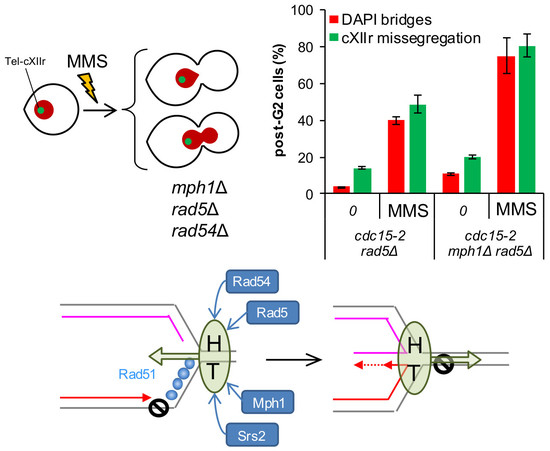
3.5. The Rad5 Helicase Prevents Aberrant post-G2 Figures upon Low Replication Stress and Mph1 Backs Up These Rad5 Safeguard Functions
The stronger MMS-dependent G2 arrest in
mph1Δ, the lack of cumulative aberrant post-G2 figures in double mutants with 3′-5′ helicases, together with the synergistic post-G2 interaction with
rad54Δ, led us to hypothesise that Mph1’s main role may lay at the beginning of the tolerance pathways that deal with replicative stress. In fact, Mph1 and its orthologs have been implicated in the regression of the SRF [
32,
33], as it has been also shown for the human Rad54 homolog [
34]. This mechanism is used to bypass the lesion by allowing DNA synthesis using the nascent lagging strand as a template for the leading strand, once this strand is left behind by the blockage. The regressed fork can then be either deregressed or process into other structures that are metabolised by the HR pathway [
5,
35,
36]. Rad5 and its human tumour-suppressor ortholog HLTF, have been implicated in fork regression as well [
35,
37,
38]. We therefore studied the cell cycle MMS profiles of
cdc15-2 rad5Δ and
cdc15-2 mph1Δ
rad5Δ. Outstandingly,
cdc15-2 rad5Δ behaved like
cdc15-2 rad51Δ and
cdc15-2 rad54Δ, yet the post-G2 aberrant phenotypes were even stronger (
Figure 6A,B). Likewise, most post-G2 abnormalities comprised bow-tie nuclei. Combination of
rad5Δ and
mph1Δ caused an additive effect; i.e., more post-G2 figures at the MMS borderline concentration. Long-term MMS sensitivity of
rad5Δ and
mph1Δ
rad5Δ phenocopied those of
rad51Δ and
rad54Δ combinations, including their epistatic relationships (
Figure 6C). We conclude that Rad5 plays an important role for a successful tolerance to replication stress, with Mph1 backing up Rad5 for a safe G2/M transition.
4. Discussion
In this work, we have characterised the cell cycle response and the post-G2 (anaphase) phenotypes that replication stress by MMS cause in a set of mutants for the HR pathway. Importantly, we have tested not a single MMS concentration but multiple scenarios that cover up to 4 orders of magnitude for MMS concentrations. This approach has let us overcome the caveat of having different G2 checkpoint sensitivities to MMS among the studied mutants. We have thus defined G2/post-G2 borderline concentrations for each mutant combination and further characterised the putative post-G2 aberrations our strains are designed for; i.e., gross chromatin anaphase bridges (DAPI bridges) and nondisjunction of the segregation-challenging chromosome XII right arm (cXIIr). The borderline parameter is almost equivalent to the EC
50 used in pharmacology and is likely the most valid approach to compare the post-G2 figures among mutants with dissimilar sensitivity to trigger and/or sustain the G2 checkpoints. In addition, the broad range of MMS concentrations allows to compare the effect of low and high replication stress since differences between these regimes, in terms of genetic pathways required for damage tolerance and survival, have been raised in recent years [
39,
40].
Although we have mainly focused on strains deficient for the Mph1 helicase (ortholog of FANCM in humans), we have also characterised several single knockout mutants for HR players during the analysis of Mph1’s genetic interactions. Just by comparing their MMS dose-response profiles, we can group our single mutants in the following categories (
Figure 7A). First, mutants that behave like that the reference “wild type” strain; i.e., only being able to trigger a sustained G2 block at 0.01% v/v MMS, with a corresponding borderline at 0.004% v/v and with a low baseline of DAPI bridges (<10%) and cXIIr missegregation (<20%):
srs2Δ. Second, mutants that differ from the former only in the abnormal post-G2 figures:
sgs1Δ and
mms4Δ. Third, mutants that yielded a moderate G2 block profile, with G2/post-G2 borderlines around 0.001% v/v, and no abnormal post-G2 figures:
mph1Δ. Fourth, mutants that yielded a much stronger G2 block profile, with G2/post-G2 borderlines at or below 0.0004% v/v, and with significant bow-tie DAPI bridges at the borderline:
rad5Δ,
rad51Δ and
rad54Δ. The bow-tie DAPI bridge is a phenotype that seems to reside in the transition between G2 and anaphase. Bow-tie nuclei get enriched after DSB generation in incomplete mutants for the G2 DNA damage checkpoint (e.g.,
rad53Δ and
chk1Δ), and are reported to be able to dynamically go back to a G2 state (mononucleated dumbbell cell) [
41]. However, similar bow-tie phenotypes are seen in
cdc15-2 top2-4, known to enter anaphase with catenations that tightly linked sister chromatids [
24]. It is worth noting that this particular phenotype was present in those mutants that show the highest sensitivity to MMS, both in the G2 arrest profiles and in the long-term survival. Furthermore, it was dependent on the MMS concentration, as there was a shift between the bow-tie nucleus and a proper G2 block as MMS concentration went higher. The best explanation for this direct relationship is that the increasing amount of MMS-driven sister chromatid linkages would result in tighter anaphases that cytologically appear as bow-tie.
Regarding the cell biology phenotypes of the
mph1Δ double mutants, we must highlight that most genetic interactions were epistatic. With respect to its contribution in the activation of and later recovery from the G2 checkpoints, these interactions posit Mph1 in linear pathways wherein Mph1 lays downstream of Rad51, Rad54 and Rad5, while laying upstream of Sgs1. However, post-G2 segregation figures surprisingly position Mph1 out of the Sgs1 pathway. To fit the observed phenotypes and Mph1’s genetic interactions in the context of the current models of DNA damage tolerance (DDT) to replication stress is challenging since the molecular mechanisms underlying DDT are far from being understood. Nevertheless, we can speculate about the roles of the different players based on the results we present here and previous literature. The novel key point is that the most evident Mph1 function takes place at early stages (presynaptic) of the HR-driven error-free DDT subpathways. The non-epistatic interactions with Rad5 and Rad54, additive in the persistence of the G2 block and synergistic in the post-G2 profiles (
Figure 2,
Figure 3,
Figure 6 and
Figure S1 and
Table 1), lead us to place Mph1 in the of RF regression subpathway (
Figure 7B). This subpathway is proposed to operate when the leading strand gets blocked by a barrier (e.g., an MMS-derived DNA adduct) [
35,
36]. Lagging strand would continue to grow in length and, as a result, ssDNA would arise in front the blocked leading strand. This ssDNA, immediately coated by the RPA complex, would trigger the G2 checkpoint. Next, the RPA-ssDNA would turn into the ssDNA-Rad51 filament and this would prepare the SRF for fork regression and annealing of the nascent lagging and leading strands. Fork regression would be possible by the helicase and/or translocase activities of Mph1, Rad5 or Rad54 [
33,
37]. Even though fork regression activity has not been demonstrated for yeast Rad54, its human homolog RAD54 owns such activity in partnership with hRAD51 [
34], supporting the inclusion of Rad54 in this model. The synergistic post-G2 profiles of
mph1Δ with
rad5Δ and
rad54Δ, would fit in this model provided that Mph1 backs up either helicase in the fork regression activity. In addition, Mph1 could collaborate with Srs2 in removing Rad51 to favour maturation of the regressed RF for DNA synthesis. This would explain the negative synergism of
mph1Δ
srs2Δ, with G2 block and long-term fitness profiles seemingly similar to the
rad mutants. It is also plausible that these helicases/translocases deregress the regressed RF [
38], bypassing the MMS abduct, and turning off the G2 checkpoint. Lastly but not least, our results pinpoint a backup role of Mph1, especially in relation to Rad5 and Rad54. Thus, the
mph1Δ mutant is intermediate in the G2 block profile and only slightly more long-term hypersensitive than the wild type (WT). We propose that both Rad54 and Rad5 lead the HR-driven DDT to MMS. To fit in this model the TLS-dependent hypermutagenic phenotypes of
mph1Δ [
21], we also propose that RF regression (or any other HR-driven subpathway) is attempted before committing cells to bypass the SRF through TLS. In the absence of Mph1, some SRFs cannot be regressed on schedule, or lead to problematic intermediates, and are diverted to be bypassed through TLS. However, in the absence of Rad5, and other presynaptic HR players, SRFs cannot be processed correctly, not even diverted towards TLS, maybe because they end up collapsing and breaking apart. Of course, the overall picture is probably more complicated since the model will need to accommodate the contribution of these players in the fate of the ssDNA gaps left behind upon blockage of the lagging strand as well as alternative mechanisms to bypass SRFs, including those that envision D-loop structures ahead of the SRF [
36].
Finally, it is intriguing that only
mms4Δ,
mms4Δ
yen1Δ and
sgs1Δ gave rise to enhanced cXIIr missegregation profiles; i.e., an increase in the ratio between cXIIr missegregation and DAPI bridges (~4:1 in these mutants versus <2:1 in the other mutants and WT). This enhancement suggests that replication stress is bypassed at the rDNA through more canonical HR mechanisms, such as template switching that results in dHJs. In this context, it is worth mentioning that RF regression might be repressed within this repetitive locus [
42,
43].
In conclusion, in this work we demonstrate that the helicase Mph1 (FANCM) contributes to the short-term tolerance to replication stress. We conclude that Mph1 deficiency leads to an extended G2 block upon replication stress and more aberrant post-G2 segregation figures when combined with deficiencies for related helicases/translocases such as Rad5 and Rad54. We believe that the plethora of short-term interactions we show here provide interesting insights for future combined interventions in the new era of personalised anti-cancer chemotherapy.