Electrode Microbial Communities Associated with Electron Donor Source Types in a Bioelectrochemical System Treating Azo-Dye Wastewater
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reactor Configuration and Operation
2.2. Chemicals and Analytical Method
2.3. Biofilm Sampling and High-Throughput 16S rRNA Gene Illumina MiSeq Sequencing
3. Results and Discussion
3.1. Performance of BESs Served with Different Electron Donor Source
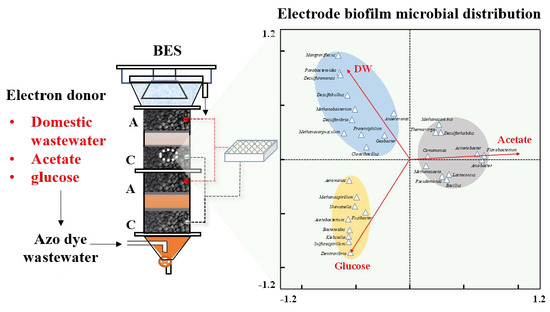
3.2. Overall Microbial Community Structures
3.3. Microbial Community Structures at Phylum, Class, and Family Levels
3.4. Potential Function of Dominant Genera
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suprun, E.V.; Radko, S.P.; Khmeleva, S.A.; Mitkevich, V.A.; Archakov, A.I.; Makarov, A.A.; Shumyantseva, V.V. Electrochemical oxidation of amyloid-beta peptide isoforms on carbon screen printed electrodes. Electrochem. Commun. 2017, 75, 33–37. [Google Scholar] [CrossRef]
- Kong, D.; Liang, B.; Yun, H.; Cheng, H.; Ma, J.; Cui, M.; Wang, A.; Ren, N. Cathodic degradation of antibiotics: Characterization and pathway analysis. Water Res. 2015, 72, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.-H.; Liu, W.-Z.; Tang, Z.-E.; Cui, D. Recent advancements in azo dye decolorization in bio-electrochemical systems (BESs): Insights into decolorization mechanism and practical application. Water Res. 2021, 203, 117512. [Google Scholar] [CrossRef]
- Cui, M.-H.; Cui, D.; Gao, L.; Cheng, H.-Y.; Wang, A.-J. Analysis of electrode microbial communities in an up-flow bioelectrochemical system treating azo dye wastewater. Electrochim. Acta 2016, 220, 252–257. [Google Scholar] [CrossRef]
- Tang, C.-C.; Zhang, X.-Y.; Wang, R.; Wang, T.-Y.; He, Z.-W.; Wang, X.C. Calcium ions-effect on performance, growth and extracellular nature of microalgal-bacterial symbiosis system treating wastewater. Environ. Res. 2022, 207, 112228. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, M.; Dębowski, M.; Kazimierowicz, J. Microwave Radiation Influence on Dairy Waste Anaerobic Digestion in a Multi-Section Hybrid Anaerobic Reactor (M-SHAR). Processes 2021, 9, 1772. [Google Scholar] [CrossRef]
- Westerholm, M.; Crauwels, S.; Van Geel, M.; Dewil, R.; Lievens, B.; Appels, L. Microwave and ultrasound pre-treatments influence microbial community structure and digester performance in anaerobic digestion of waste activated sludge. Appl. Microbiol. Biotechnol. 2016, 100, 5339–5352. [Google Scholar] [CrossRef]
- Dębowski, M.; Zieliński, M.; Kisielewska, M.; Kazimierowicz, J. Evaluation of Anaerobic Digestion of Dairy Wastewater in an Innovative Multi-Section Horizontal Flow Reactor. Energies 2020, 13, 2392. [Google Scholar] [CrossRef]
- Lyu, W.; Song, Q.; Shi, J.; Wang, H.; Wang, B.; Hu, X. Weak magnetic field affected microbial communities and function in the A/O/A sequencing batch reactors for enhanced aerobic granulation. Sep. Purif. Technol. 2021, 266, 118537. [Google Scholar] [CrossRef]
- Zhao, B.; Sha, H.; Li, J.; Cao, S.; Wang, G.; Yang, Y. Static magnetic field enhanced methane production via stimulating the growth and composition of microbial community. J. Clean. Prod. 2020, 271, 122664. [Google Scholar] [CrossRef]
- Daghio, M.; Gandolfi, I.; Bestetti, G.; Franzetti, A.; Guerrini, E.; Cristiani, P. Anodic and cathodic microbial communities in single chamber microbial fuel cells. New Biotechnol. 2015, 32, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.; Ji, M.; Zhao, Y.; Pavlostathis, S.G.; Zhao, Q. Effects of salinity and COD/N on denitrification and bacterial community in dicyclic-type electrode based biofilm reactor. Chemosphere 2017, 192, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yates, M.D.; Hatzell, M.C.; Rao, H.A.; Saikaly, P.E.; Logan, B.E. Microbial Community Composition Is Unaffected by Anode Potential. Environ. Sci. Technol. 2013, 48, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.-H.; Cui, D.; Gao, L.; Wang, A.-J.; Cheng, H.-Y. Azo dye decolorization in an up-flow bioelectrochemical reactor with domestic wastewater as a cost-effective yet highly efficient electron donor source. Water Res. 2016, 105, 520–526. [Google Scholar] [CrossRef]
- Mu, Y.; Rabaey, K.; Rozendal, R.A.; Yuan, Z.; Keller, J. Decolorization of Azo Dyes in Bioelectrochemical Systems. Environ. Sci. Technol. 2009, 43, 5137–5143. [Google Scholar] [CrossRef]
- Cheng, H.-Y.; Liang, B.; Mu, Y.; Cui, M.-H.; Li, K.; Wu, W.; Wang, A.-J. Stimulation of oxygen to bioanode for energy recovery from recalcitrant organic matter aniline in microbial fuel cells (MFCs). Water Res. 2015, 81, 72–83. [Google Scholar] [CrossRef]
- Quan, X.-C.; Quan, Y.-P.; Tao, K. Effect of anode aeration on the performance and microbial community of an air–cathode microbial fuel cell. Chem. Eng. J. 2012, 210, 150–156. [Google Scholar] [CrossRef]
- Liang, B.; Cheng, H.; Van Nostrand, J.; Ma, J.; Yu, H.; Kong, D.; Liu, W.; Ren, N.; Wu, L.; Wang, A.; et al. Microbial community structure and function of Nitrobenzene reduction biocathode in response to carbon source switchover. Water Res. 2014, 54, 137–148. [Google Scholar] [CrossRef]
- Cui, M.-H.; Cui, D.; Lee, H.-S.; Liang, B.; Wang, A.-J.; Cheng, H.-Y. Effect of electrode position on azo dye removal in an up-flow hybrid anaerobic digestion reactor with built-in bioelectrochemical system. Sci. Rep. 2016, 6, 25223. [Google Scholar] [CrossRef] [Green Version]
- Bond, D.R.; Lovley, D.R. Electricity Production by Geobacter sulfurreducens Attached to Electrodes. Appl. Environ. Microbiol. 2003, 69, 1548–1555. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Zhou, J.; Chen, C.; Wang, J.; Jin, R.; Lv, H. Decolorization of azo dyes by Geobacter metallireducens. Appl. Microbiol. Biotechnol. 2012, 97, 7935–7942. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.S.; Eaktasang, N.; Kwon, D.-Y.; Kim, H.S. Enhanced current production by Desulfovibrio desulfuricans biofilm in a mediator-less microbial fuel cell. Bioresour. Technol. 2014, 165, 27–30. [Google Scholar] [CrossRef]
- Kim, S.-Y.; An, J.-Y.; Kim, B.-W. Improvement of the decolorization of azo dye by anaerobic sludge bioaugmented with Desulfovibrio desulfuricans. Biotechnol. Bioprocess Eng. 2007, 12, 222–227. [Google Scholar] [CrossRef]
- Sass, H.; Ramamoorthy, S.; Yarwood, C.; Langner, H.; Schumann, P.; Kroppenstedt, R.M.; Spring, S.; Rosenzweig, R.F. Desulfovibrio idahonensis sp. nov., sulfate-reducing bacteria isolated from a metal(loid)-contaminated freshwater sediment. Int. J. Syst. Evol. Microbiol. 2009, 59, 2208–2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhang, D.; Huang, X.; Qin, W. Acinetobacter harbinensis sp. nov., isolated from river water. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 5, 1507–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; Zhang, W.; Ma, J.; Cai, J.; Li, S.; Zhu, X.; Yang, G.; Zhao, X. Biodegradation of Azo Dye Disperse Orange S-RL by a Newly Isolated Strain Acinetobacter sp. SRL8. Water Environ. Res. 2015, 87, 516–523. [Google Scholar] [CrossRef]
- Pham, T.H.; Boon, N.; De Maeyer, K.; Höfte, M.; Rabaey, K.; Verstraete, W. Use of Pseudomonas species producing phenazine-based metabolites in the anodes of microbial fuel cells to improve electricity generation. Appl. Microbiol. Biotechnol. 2008, 80, 985–993. [Google Scholar] [CrossRef]
- Qiao, Y.; Qiao, Y.-J.; Zou, L.; Ma, C.-X.; Liu, J. Real-time monitoring of phenazines excretion in Pseudomonas aeruginosa microbial fuel cell anode using cavity microelectrodes. Bioresour. Technol. 2015, 198, 1–6. [Google Scholar] [CrossRef]
- Vandamme, P.; Falsen, E.; Rossau, R.; Hoste, B.; Segers, P.; Tytgat, R.; De Ley, J. Revision of Campylobacter, Helicobacter, and Wolinella Taxonomy: Emendation of Generic Descriptions and Proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 1991, 41, 88–103. [Google Scholar] [CrossRef] [Green Version]
- Toh, H.; Sharma, V.K.; Oshima, K.; Kondo, S.; Hattori, M.; Ward, F.B.; Free, A.; Taylor, T.D. Complete Genome Sequences of Arcobacter butzleri ED-1 and Arcobacter sp. Strain L, Both Isolated from a Microbial Fuel Cell. J. Bacteriol. 2011, 193, 6411–6412. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-Y.; Kim, T.G.; Cho, K.-S. Enhancement of electricity production in a mediatorless air–cathode microbial fuel cell using Klebsiella sp. IR21. Bioprocess Biosyst. Eng. 2016, 39, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Elizalde-González, M.; Fuentes, L.; Guevara-Villa, M. Degradation of immobilized azo dyes by Klebsiella sp. UAP-b5 isolated from maize bioadsorbent. J. Hazard. Mater. 2009, 161, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Balch, W.E.; Schoberth, S.; Tanner, R.S.; Wolfe, R.S. Acetobacterium, a New Genus of Hydrogen-Oxidizing, Carbon Dioxide-Reducing, Anaerobic Bacteria. Int. J. Syst. Bacteriol. 1977, 27, 355–361. [Google Scholar] [CrossRef] [Green Version]
- Ma, K. Methanosaeta harundinacea sp. nov., a novel acetate-scavenging methanogen isolated from a UASB reactor. Int. J. Syst. Evol. Microbiol. 2006, 56 Pt 1, 127–131. [Google Scholar] [CrossRef]
- Liu, F.; Rotaru, A.-E.; Shrestha, P.M.; Malvankar, N.S.; Nevin, K.P.; Lovley, D.R. Promoting direct interspecies electron transfer with activated carbon. Energy Environ. Sci. 2012, 5, 8982–8989. [Google Scholar] [CrossRef] [Green Version]
- Rotaru, A.-E.; Shrestha, P.M.; Liu, F.; Shrestha, M.; Shrestha, D.; Embree, M.; Zengler, K.; Wardman, C.; Nevin, K.P.; Lovley, D.R. A new model for electron flow during anaerobic digestion: Direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ. Sci. 2013, 7, 408–415. [Google Scholar] [CrossRef]
- Han, R.; Yuan, Y.; Cao, Q.; Li, Q.; Chen, L.; Zhu, D.; Liu, D. PCR–DGGE Analysis on Microbial Community Structure of Rural Household Biogas Digesters in Qinghai Plateau. Curr. Microbiol. 2017, 75, 541–549. [Google Scholar] [CrossRef]
- Tan, H.-Q.; Li, T.-T.; Zhu, C.; Zhang, X.-Q.; Wu, M.; Zhu, X.-F. Parabacteroides chartae sp. nov., an obligately anaerobic species from wastewater of a paper mill. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 11, 2613–2617. [Google Scholar] [CrossRef]
- Coates, J.D.; Lonergan, D.J.; Philips, E.J.P.; Jenter, H.; Lovley, D.R. Desulfuromonas palmitatis sp. nov., a marine dissimilatory Fe(III) reducer that can oxidize long-chain fatty acids. Arch. Microbiol. 1995, 164, 406–413. [Google Scholar] [CrossRef]
- Krumholz, L.R. Desulfuromonas chloroethenica sp. nov. Uses Tetrachloroethylene and Trichloroethylene as Electron Acceptors. Int. J. Syst. Bacteriol. 1997, 47, 1262–1263. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Cao, X.-X.; Liang, P.; Huang, X.; Yang, S.-P.; Zhao, G.-G. Electricity generation from glucose by a Klebsiella sp. in microbial fuel cells. Appl. Microbiol. Biotechnol. 2010, 87, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-Y.; Zhen, S.-H.; Chao, S.-L.; Wu, J.; Cheng, L.; Li, S.-W.; Xiao, X.; Zhou, X. Electrochemistry of newly isolated Gram-positive bacteria Paenibacillus lautus with starch as sole carbon source. Electrochim. Acta 2022, 411, 140068. [Google Scholar] [CrossRef]
- Mai, Q.; Yang, G.; Cao, J.; Zhang, X.; Zhuang, L. Stratified microbial structure and activity within anode biofilm during electrochemically assisted brewery wastewater treatment. Biotechnol. Bioeng. 2020, 117, 2023–2031. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pan, Y.; Zhu, T.; Wang, A.; Lu, Y.; Lv, L.; Zhang, K.; Li, Z. Enhanced performance and microbial community analysis of bioelectrochemical system integrated with bio-contact oxidation reactor for treatment of wastewater containing azo dye. Sci. Total Environ. 2018, 634, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-H.; Cheng, H.-Y.; Ding, Y.-C.; Su, S.-G.; Wang, B.; Zeng, R.; Sharif, H.M.A.; Wang, A.-J. Enhanced treatment of coal gasification wastewater in a membraneless sleeve-type bioelectrochemical system. Bioelectrochemistry 2019, 129, 154–161. [Google Scholar] [CrossRef]






| Mark | Sample Name | Number of Sequences | OTU | Chao 1 | ACE | Shannon | Simpson |
|---|---|---|---|---|---|---|---|
| C1 | RDW down-cathode | 29,514 | 649 | 699 | 715 | 4.29 | 0.04 |
| C2 | RDW down-anode | 28,786 | 625 | 662 | 682 | 4.23 | 0.04 |
| C3 | RDW up-cathode | 31,281 | 539 | 592 | 609 | 3.67 | 0.07 |
| C4 | RDW up-anode | 30,224 | 577 | 629 | 644 | 3.79 | 0.06 |
| C5 | RAc down-cathode | 33,033 | 335 | 356 | 373 | 2.80 | 0.17 |
| C6 | RAc down-anode | 34,760 | 346 | 400 | 408 | 3.10 | 0.11 |
| C7 | RAc up-cathode | 35,182 | 398 | 468 | 455 | 3.12 | 0.15 |
| C8 | RAc up-anode | 33,810 | 351 | 388 | 415 | 3.30 | 0.08 |
| C9 | RGlu down-cathode | 32,182 | 376 | 421 | 438 | 3.23 | 0.13 |
| C10 | RGlu down-anode | 30,776 | 387 | 464 | 469 | 3.63 | 0.06 |
| C11 | RGlu up-cathode | 31,687 | 354 | 383 | 393 | 3.30 | 0.12 |
| C12 | RGlu up-anode | 31,257 | 402 | 463 | 483 | 3.29 | 0.10 |
| Genus | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | C11 | C12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methanosaeta | 0.70 | 0.27 | 19.00 | 16.46 | 37.25 | 26.26 | 35.83 | 15.43 | 17.29 | 4.84 | 18.94 | 18.51 |
| Acinetobacter | 1.53 | 1.67 | 1.68 | 2.03 | 18.76 | 32.89 | 18.26 | 38.51 | 0.74 | 2.57 | 1.38 | 2.50 |
| Pseudomonas | 0.55 | 0.48 | 0.51 | 0.86 | 2.05 | 7.80 | 5.21 | 8.35 | 0.59 | 4.99 | 1.46 | 3.54 |
| Geobacter | 1.65 | 14.15 | 0.46 | 11.84 | 1.71 | 8.35 | 0.35 | 6.75 | 0.26 | 8.02 | 0.12 | 7.68 |
| Arcobacter | 0.04 | 0.08 | 0.04 | 0.08 | 1.09 | 5.14 | 1.23 | 8.76 | 0.07 | 0.75 | 0.11 | 0.32 |
| Thermovirga | 0.16 | 0.03 | 2.16 | 1.86 | 6.34 | 0.16 | 1.98 | 0.29 | 0.47 | 0.09 | 0.52 | 0.07 |
| Desulforhabdus | 0.19 | 0.32 | 1.49 | 1.73 | 4.90 | 0.76 | 2.70 | 0.28 | 0.31 | 0.01 | 0.39 | 0.14 |
| Desulfovibrio | 13.96 | 11.99 | 6.30 | 8.31 | 1.88 | 0.16 | 2.00 | 0.26 | 4.71 | 4.84 | 6.50 | 2.23 |
| Desulfobulbus | 4.34 | 3.82 | 0.15 | 2.16 | 0.29 | 0.07 | 0.18 | 0.07 | 1.00 | 0.55 | 0.51 | 0.36 |
| Methanobacterium | 0.26 | 0.04 | 8.07 | 0.03 | 0.05 | 0.02 | 1.04 | 0.04 | 0.43 | 0.07 | 1.81 | 0.45 |
| Parabacteroides | 2.19 | 3.51 | 0.88 | 1.32 | - | 0.02 | 0.01 | 0.01 | 0.15 | 0.36 | 0.10 | 0.13 |
| Desulfuromonas | 0.12 | 3.93 | 0.36 | 2.90 | 0.02 | 0.00 | 0.01 | 0.04 | 0.48 | 0.05 | 0.24 | 0.01 |
| Klebsiella | 1.24 | 0.82 | 1.07 | 0.78 | 0.02 | 0.04 | 0.04 | 0.01 | 3.24 | 17.43 | 2.44 | 15.33 |
| Acetobacterium | 1.30 | 1.65 | 0.17 | 0.51 | - | - | 0.00 | - | 4.87 | 7.27 | 3.28 | 1.22 |
| Aeromonas | 2.52 | 1.04 | 0.82 | 0.69 | 0.02 | 0.21 | 0.05 | 0.13 | 0.89 | 4.03 | 0.62 | 3.16 |
| Denitrovibrio | - | 0.02 | - | 0.02 | 0.01 | 0.03 | 0.01 | 0.02 | 0.04 | 5.94 | 0.01 | 1.01 |
| Fusibacter | 0.31 | 0.54 | 0.29 | 0.20 | 0.04 | 0.70 | 0.17 | 0.21 | 0.08 | 3.41 | 0.12 | 2.94 |
| Proteiniphilum | 1.00 | 0.52 | 2.35 | 1.86 | 0.30 | 0.43 | 0.48 | 0.13 | 1.01 | 0.18 | 2.22 | 0.17 |
| Cloacibacillus | 1.40 | 1.18 | 1.04 | 0.57 | 0.19 | 0.32 | 0.71 | 0.20 | 1.17 | 0.16 | 1.91 | 0.23 |
| Mangroviflexus | 2.58 | 0.39 | 0.10 | 0.06 | - | - | - | - | - | 0.01 | - | - |
| Anaerovorax | 1.22 | 0.56 | 0.73 | 0.51 | 0.52 | 0.38 | 0.60 | 0.48 | 0.23 | 0.19 | 0.14 | 0.12 |
| Methanocorpusculum | 2.02 | 0.29 | - | 0.01 | 0.01 | 0.00 | 0.04 | 0.00 | 1.12 | 0.10 | 0.18 | 0.09 |
| Methanosarcina | 1.05 | 0.20 | 0.17 | 0.63 | 2.76 | 0.49 | 0.29 | 0.56 | 0.06 | 0.01 | 0.10 | 0.02 |
| Shewanella | 0.53 | 0.50 | 0.32 | 0.22 | 0.03 | 0.14 | 0.09 | 0.13 | 0.80 | 2.38 | 0.62 | 1.50 |
| Comamonas | 0.31 | 0.43 | 0.13 | 0.31 | 0.32 | 0.56 | 0.26 | 2.08 | 0.08 | 0.49 | 0.11 | 0.65 |
| Methanospirillum | 0.24 | 0.01 | 0.89 | 0.00 | 0.04 | 0.01 | 0.25 | 0.00 | 1.82 | 0.19 | 0.97 | 0.06 |
| Lactococcus | 0.09 | 0.07 | 0.10 | 0.15 | 0.47 | 1.44 | 2.11 | 1.62 | 0.14 | 0.41 | 0.60 | 0.93 |
| Bacteroides | 0.10 | 0.15 | 0.03 | 0.03 | - | 0.00 | 0.02 | 0.01 | 0.29 | 1.16 | 0.21 | 0.65 |
| Bacillus | 0.04 | 0.05 | 0.04 | 0.13 | 0.42 | 1.31 | 2.00 | 1.44 | 0.06 | 0.34 | 0.79 | 1.03 |
| Sulfurospirillum | 0.07 | 0.02 | 0.02 | 0.01 | 0.00 | 0.00 | - | - | 0.03 | 1.59 | 0.01 | 0.09 |
| Flavobacterium | 0.04 | 0.03 | 0.01 | 0.03 | 0.12 | 0.69 | 0.33 | 2.89 | 0.02 | 0.08 | 0.03 | 0.06 |
| Unclassified | 49.70 | 44.25 | 43.67 | 36.64 | 15.93 | 8.01 | 18.17 | 7.63 | 49.76 | 21.83 | 47.64 | 28.93 |
| Others # (<1%) | 8.53 | 6.99 | 6.95 | 7.09 | 4.48 | 3.59 | 5.58 | 3.66 | 7.80 | 5.69 | 5.89 | 5.85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.; Zhang, L.; Cui, M.-H.; Wang, A. Electrode Microbial Communities Associated with Electron Donor Source Types in a Bioelectrochemical System Treating Azo-Dye Wastewater. Water 2022, 14, 1505. https://0-doi-org.brum.beds.ac.uk/10.3390/w14091505
Guo Z, Zhang L, Cui M-H, Wang A. Electrode Microbial Communities Associated with Electron Donor Source Types in a Bioelectrochemical System Treating Azo-Dye Wastewater. Water. 2022; 14(9):1505. https://0-doi-org.brum.beds.ac.uk/10.3390/w14091505
Chicago/Turabian StyleGuo, Zechong, Lu Zhang, Min-Hua Cui, and Aijie Wang. 2022. "Electrode Microbial Communities Associated with Electron Donor Source Types in a Bioelectrochemical System Treating Azo-Dye Wastewater" Water 14, no. 9: 1505. https://0-doi-org.brum.beds.ac.uk/10.3390/w14091505