1. Introduction
1.1. Broader context: the progression of information optimization phase transitions
One way to view the history of the universe is as a continuous evolution of nodes of information optimization. Each subsequent node is able to optimize information better, faster, and at a higher order of complexity than the last, and improve itself by evolving more quickly. Each previous node is required for the next node. Subsequent nodes eventually emerge as distinct, but may overlap. Earlier nodes may continue to coexist with subsequent nodes. Information optimization has a cyclic nature, developing in environments of symmetry and simplicity, then continuously breaking symmetry and evolving through instability to reorganize into increasingly dense nodes of complexity and diversity.
Perhaps the most obvious progression of information optimization begins with the formation of the universe and extends to contemporary human intelligence. Some of the earliest nodes could include atoms, matter, and amino acids. Eventually, the chemical properties of matter evolved into a second node, biology, DNA, and self-replication. Natural selection developed as a third node of information optimization, being a higher order of random combination. Then intelligence, much quicker than natural selection, evolved as the fourth node. Intelligence created the fifth node, technology. Technology is able to optimize information better, faster, and at a higher order of complexity than humans in some cases, and could eventually challenge human intelligence. The sixth node is next, locally a phase transition in intelligence, and globally one of the ongoing phase transitions in information optimization.
1.2. Symmetry-breaking and present instability
In attempting to self-organize for a phase transition in intelligence, it could be argued that the progression of information optimization is currently out of balance and unstable. A contemporary issue has arisen that given its exponential growth, technology could be a potential sole successor to human intelligence. The trajectory of supercomputing exemplifies this instability (
Figure 1). As of November 2009, the world’s fastest supercomputer was the Cray Jaguar located at the U.S. Department of Energy’s Oak Ridge National Laboratory, operating at 1.8 petaflops (1.8 x 10
15 flops) [
1]. Unlike human brain capacity, supercomputing capacity has been growing exponentially. In June 2005, the world’s fastest supercomputer was the IBM Blue Gene/L at Los Alamos National Laboratory, running at 0.1 petaflops [
2]. In less than five years, the Jaguar represents an order of magnitude increase, the latest culmination of capacity doublings each few years.
The next supercomputing node, one more order of magnitude, at 10
16 flops, is expected in 2011 with the Pleiades, Blue Waters, or Japanese RIKEN systems. 10
16 flops would possibly allow the functional simulation of the human brain. Clearly, there are many critical differences between the human brain and supercomputers. Supercomputers tend to be modular in architecture and address specific problems as opposed to having the general problem solving capabilities of the human brain. Having equal to or greater than human-level raw computing power in a machine does not necessarily confer the ability to compute as a human. Some estimates of the raw computational power of the human brain range between 10
13 and 10
16 operations per second [
4]. This would indicate that supercomputing power is already on the order of estimated human brain capacity, but intelligent or human-simulating machines do not yet exist. The digital comparison of raw computational capability may not be the right measure for understanding the complexity of the brain. Signal transmission is different in biological systems, with a variety of parameters such as context and continuum determining the quality and quantity of signals.
The architecture of distributed computing might allow problems to be processed in a fashion more similar to that of the human brain. The largest current project in distributed computing, Stanford protein Folding@home, reached 5 petaflops (5 x 10
15 flops) in computing capacity in early 2009, just as supercomputers were reaching 1 petaflop [
5]. It is likely that there will be many steps in fully emulating human neural processes. Of equal salience is that this is not some distant point in the future but already a contemporary research challenge. Another big leap in supercomputing growth could be with three more orders of magnitude, at 10
19 flops, expected in 2018, which might allow full human brain neural simulation.
1.3. Engineering life into technology: the complex dynamical response
One response to the imbalance and uncertainty regarding the future of intelligence is an emerging complex dynamical system, the engineering of life into technology. Engineering life into technology could potentially reassert symmetry and eventually bring about a phase transition in intelligence. This could happen through the mastery and augmentation of all biological processes, and the unification of organic and inorganic matter.
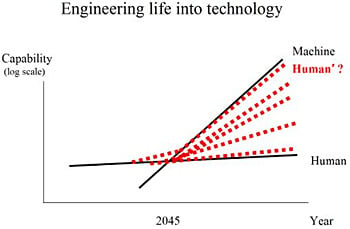
Figure 2 is a simple representation of the imbalance and some possible solutions. The growth in the capability of machines and humans is shown over time. Machine capacity obviously grows faster than human capacity. On current trajectories, a crossover point would be inevitable, and could possibly occur within the next few decades. Some estimates for the timing of the crossover point include 2045 per futurist Ray Kurzweil [
6], and possibly before 2040 per mathematician Vernor Vinge [
7]. Specifically, this is the moment where machines might have greater than human-level intelligence, sometimes referred to as the technological singularity. Traditionally, life and technology have been thought of as disparate. One path forward is the emerging complex system described here, the engineering of life into technology, that can possibly keep pace with technological advances (red-dotted Human’ line). The long-term future of intelligence could include a variety of different mergers and integrations of life and technology (red-dotted lines).
1.4. Description of methods and scope of analysis
The engineering of life into technology is already occurring at many levels and could start to be recognized as a pervasive phenomenon. This analysis provides a comprehensive identification and examination of diverse areas where this is happening. Specifically, recent high-impact research advances from a variety of fields are described in straightforward terms, and how complexity theory may be applied to understand how these advances together could trigger a phase transition in intelligence. Information was gathered from technical conferences, scientific publications, and informal interviews with researchers in multiple fields from February to November 2009.
Figure 3 depicts the simultaneously occurring macroscopic and microscopic phenomena that comprise the engineering of life into technology. Four classes of macroscopic themes are represented: conceptual shifts, the fundamentals of DNA, biomedical applications, and intelligence. Within these macroscopic themes, there are numerous microscopic elements acting as independent agents in the complex dynamical system. These include first, the evolving notions of science, the scientific method, health, and preventive medicine; second, sequencing and synthesizing DNA, and characterizing the whole contents of cells; third, optimizing biology through nanomedicine, surgical intervention, regenerative medicine, and longevity therapies; and fourth, extending intelligence through neuroscience, artificial intelligence and robotics, whole brain simulation, and human-machine integration interfaces.
The scope of this analysis focuses primarily on the current status of technologies that may be self-organizing for a potential phase transition in intelligence. There are certainly many other potential influences on technology development and deployment such as government regulation, public policy, legal frameworks, economic motivations, and social acceptance that are not considered here. A second point of acknowledgment is that many of the described technologies are forward-looking and speculative. There are a myriad of privacy, security, fair use, accessibility, right to decline adoption, and other issues regarding nearly all of the technologies that are also not considered here. One example would be mechanisms for ensuring privacy and non-discrimination in an era of low-cost whole human genome sequencing, such as labs requiring a signed affidavit as is currently under discussion in the U.K. Finally, it should be noted that many of the described technologies are in early stages of development, have valid opposing criticisms, and may not ever come to fruition, all of which would be expected in a complex dynamical systems environment with competing and evolving elements.
2. Conceptual shifts
The first macroscopic phenomenon in the process of engineering life into technology is several shifts at the conceptual level that are starting to transform overall thinking about science and health.
2.1. Science and the scientific method
The notion of what constitutes science and the scientific method has been evolving. What once may have been perceived as an art has become an information technology and engineering problem. Traditional characterization and experimentation activities are being supplemented with two new steps in a virtuous feedback loop. First, mathematical modeling and computer simulation have been added to define and predict behavior using platforms such as SimTK, Bio-SPICE, BioCad, and Entelos. Second, where possible, scientists may be engineering and building live organisms or other examples in the lab to demonstrate mastery and develop new uses [
8]. An indication of the increasing centrality of computing is the effort to include computational thinking principles such as logic, recursion, abstraction, and program design as a cornerstone skill for all students, commensurate with reading, writing, and arithmetic [
9]. An intelligent person must be able to think and express ideas in the medium of the day. Computer scientist and physicist Stephen Wolfram has gone even further to suggest that the science and technology of the future may be increasingly non-human understandable as the computational universe of all possible technologies is sampled and applied to human use [
10].
Another important conceptual shift that is influencing many areas, including the notions and conduct of science, is social networking. There have been several new paradigms in technology that have had a pervasive influence: the mainframe computer, the personal computer, the internet, and now, social networking. In science, social networking facilitates the identification of potential peers and the ability to collaborate. Social networking and open-source software communities have influenced the emergence of a variety of open research initiatives in science such as open access journals, open data, open notebook science, and open source drug discovery [
11].
2.2. Health and preventive medicine
A second relevant domain of conceptual change is occurring in health and medicine. One shift is the move from ‘health care’ to ‘health.’ While health care may connote a point resolution to a problem that has occurred, health suggests the ongoing monitoring and management of the quality of health.
The continuum of what may constitute health is becoming broader. Outcomes are expanding to include not just cure but also improvement, baseline normalization, prevention, enhancement, and possibly, self-expression [
8]. The focus on cures now includes both high-priority infectious disease, and chronic disease and aging. According to the World Health Organization (WHO), chronic disease is the world’s greatest killer, cardiovascular disease at present, and cancer by 2010 [
12]. Being healthy may not mean just physically, but also mentally and environmentally. In many ways aging and death are still taboo topics, for example few step forward to debate the high percent of health care costs spent in the last few months of life. However, there is starting to be some consideration of a postmortal society, probing the possibility of “deferring death, addressing its causes, altering its boundaries, and controlling all of its parameters [
13].”
While the resolution of death and aging may not be immediate, preventive medicine is already becoming an established field. In the U.S., the market for tailoring treatments to individuals is estimated to be
$225-232 billion in 2009, growing at 11% per year [
14]. The full implementation of preventive medicine could be in three tiers (
Figure 4). Automated devices and web-based tools, telediagnosis, health social networks, and self-diagnosis could be the first line of information and defense for individuals. 61% of American adults (and 83% of internet users) currently seek health information on the internet [
15]. Automated health monitoring devices could be responsible for not just collecting information pertaining to symptoms, but also assessing an individual’s ongoing health climate. The second line of defense could be a new class of health professionals known as wellness coaches or health advisors (analogous to financial advisors). These professionals would have the training and certification to integrate various health data streams (
i.e., genomics, phenotypic biomarker, family and health history, behavior, and environment) into unified, personalized health management plans. Third, traditional health care systems could be relied upon for their expertise in treating disease states, emergencies, and other exceptions.
3. Fundamentals of DNA
The conceptual broadening of science and health creates a platform for the second macroscopic phenomenon in the process of engineering life into technology, the fundamentals of DNA. A full understanding of genomics, the instruction set for life, could mean the ability to more comprehensively manipulate both the world around us and the world within us. Among other possibilities, this could allow the development of the next generation of biotechnology, not just altering one gene or element as with present techniques, but potentially creating full biological organisms in precise and reliable detail from scratch. The building blocks for this are the sequencing (reading) and synthesizing (writing) DNA.
Moore’s Law is a familiar concept from computing. Specifically, it refers to the long-term trend of being able to double the number of transistors placed on a chip approximately each eighteen months for about the same price. These price/performance doublings have been observed across nearly all areas of information processing and communications, and in computing even before transistors. There is a similar analog in life science technologies, Carlson Curves (
Figure 5). This is a logarithmic plot of the decreasing cost per base pair of sequenced and synthesized DNA. DNA sequencing has been progressing at faster than Moore’s Law rates, with capacity doublings each twelve months, and accelerating even faster in the last few years. DNA synthesis, on the other hand, has not been progressing as fast, either for oligonucleotide synthesis (shorter strands) or gene synthesis (longer strands), and is an area of present research.
3.1. DNA sequencing
A major milestone in contemporary genetics research occurred in 2003 with the completed sequencing of the human genome. $3 billion was spent to sequence the 3 billion nucleotide base pairs that make up human DNA. When stored on a computer, 3 billion base pairs take up about 3 gigabytes of memory (the size of a movie file). A surprising finding from the human genome project was that humans are relatively simple genetically, having only approximately 21,000 genes, less than some organisms. What appears to be driving the higher levels of complexity in humans is the mixing and matching of gene products.
3.1.1. Personal genome revolution
The rapidly decreasing cost of genomic sequencing suggests the potential for its widespread use in research, clinical, and consumer applications. As of November 2009, the cost of whole human genome sequencing was
$4,400 (for materials costs) for researchers [
17] and
$48,000 for consumers [
18]. Already since 2007, a variety of offerings have been available directly to consumers for
$300-
$1,100. These consumer genomic services genotype 600,000 to one million SNPs (single nucleotide polymorphisms; locations of genetic variance) and map them to 30-130 different conditions such as Alzheimer’s disease, cancers, heart attack, and drug response [
18]. The two most important current applications of personalized genomics are in identifying drug response and in routing higher risk individuals to earlier screenings for disease detection. Additionally, genomics is one of the first quantitative health data streams that can be used in the implementation of preventive medicine.
An era of low-cost genomic sequencing could allow biobanks to be created. Biobanks are large databases with potentially millions of participants contributing genomic and phenotypic biomarker data, and possibly eventually neural data. Medical research could be accelerated as pharmaceutical companies, researchers, entrepreneurs, patients, and other interested parties search these data-rich pre-aggregated communities of individuals. The long tail of medicine could be enabled with a focus on ultra-specific cohorts and orphan diseases that were unfeasible to reach previously and will likely be required in an era of personalized medicine.
3.1.2. Advances in DNA sequencing technology
Ratcheting down the Carlson Curves, advances in DNA sequencing technology are being closely watched for the advent of affordable whole human genome sequencing. Third-generation sequencing company Pacific Biosciences has announced the estimated launch of their single-molecule real-time (SMRT
TM) sequencing technology for the second half of 2010 [
19], which could enable
$100 whole human genome sequencing by 2014. A pyrosequencing technique is used, one method of sequencing by synthesis, essentially recording the activity of the DNA polymerase enzyme as it synthesizes a copy of DNA. Two key innovations may deliver a 30,000-fold improvement over current approaches [
20]. First, a fluorescent label is attached to the terminal phosphate of each nucleotide rather than the nucleotide itself to recognize the base (
Figure 6), which speeds up the process. Second, a small-volume zero-mode waveguide optical chamber was created to read the bases more efficiently.
Sequencing is likely to persist as an indispensable scientific tool for understanding the complex biological relationships of DNA, RNA, protein, and metabolite levels. Fourth-generation sequencing methods are already under development. One prominent example is Oxford Nanopore Technologies, sequencing DNA as it is pushed through a nanopore by reading the distinct electrical signatures of the bases as they bind transiently to a cyclodextrin (sugar molecule) in the pore (
Figure 6). Other examples include Halcyon Molecular and ZS Genetics who are developing ultra-fast electron microscopy-based DNA sequencing technology.
3.2. DNA synthesis
After sequencing DNA, the next important application is synthesizing it. Generally called synthetic biology or bioengineering, this field is concerned with the programming and engineering of genetic sequences for specific outputs such as pharmaceuticals, energy, and toxin sensors. DNA can be conceptualized as the ‘chemical software’ that operates living organisms. DNA sequences can be designed by computer and then printed physically with a DNA synthesizer, similar in concept to a laser printer, having separate cartridges for the four nucleotide bases (A, C, G, and T) and other binding molecules (
Figure 6).
3.2.1. Next-generation synthetic biologists
In addition to increasing numbers of synthetic biology technical conferences, bioengineering is attracting enthusiastic inventors in the form of the hundreds of college students who participate in the annual International Genetically Engineered Machines competition, iGEM, to create new biological parts or functions. In 2009, 102 teams from twenty-five countries competed. Gold-medal winners distinguished themselves in a variety of ways including by creating an original DNA-based biological part, appropriately cataloging and documenting the part, helping at least one other team, and addressing biosafety protocols [
21]. Some of the winning innovations included a graded sensing system designed to change color in response to different toxins, a biological version of an LCD screen made of yeast, and a system for constructing mammalian synthetic promoters (sites on DNA where RNA transcription begins) [
21]. Complementing professional bioengineering is the DIYbio.org community, worldwide amateur biologists who are undertaking projects such as shot-glass DNA extraction [
22] and harmful chemical detection with the melaminometer (finding melamine in contaminated milk) [
23].
3.2.2. Bioengineering tools
As with other engineering-driven disciplines such as materials and computer science, tools are critical and must be built from scratch. At present, bioengineering processes are relatively expensive and not automated or standardized. Although following Carlson Curves, the present cost for DNA synthesis is still high,
$0.50-
$1.00 per base pair, meaning that it would cost
$1.5-3 billion to synthesize a full human genome. Most of the cost is related to design and manufacture. Currently short oligonucleotide sequences (up to 200 base pairs) can be synthesized reliably, but a low-cost repeatable synthesis method for genes and genomes extending into the millions of base pairs is needed. Further, design capability lags synthesis capability, being about 400-800-fold less capable and allowing only 10,000-20,000 base pair systems to be fully forward-engineered at present [
24]. There is work in progress to develop computer-aided design tools and test platforms for synthetic biology [
25].
In addition to tools for the automated design and manufacture of DNA, another important bioengineering tool is reusable software components. Analogous to building a house, it is much easier to start with standardized lumber rather than by growing a tree. Most contemporary software engineers work at high levels of abstraction and do not concern themselves with the ones and zeros of machine language. In the future, bioengineers would not need to work directly with the Ac, Cs, Gs, and Ts of DNA, or understand the architecture of basic elements such as promoters, terminators, and open reading frames. A key industry resource is the Registry of Standard Biological Parts [
26]. The parts registry is an open source directory with 5,230 listings as of November 2009, organized into four levels of abstraction: DNA, parts, devices, and systems. These off-the-shelf building blocks encode specific biological functions in snippets of DNA that are ready to be joined modularly with other DNA parts (
Figure 6). Once a series of parts is assembled, it can be printed into physical form via a DNA synthesizer or ordered from any one of a few hundred worldwide DNA synthesis labs. Some examples of DNA parts include green fluorescent proteins, cell signaling mechanisms, and ribosome binding sites.
3.2.3. Minimal genomes
In working with DNA, a natural question arises as to the minimal genome required for life. This could be helpful when designing new applications with maximum efficiency. Some intermediary steps towards the creation of minimal genomes have been accomplished. The first synthesis of an organism’s full genome was announced in December 2008, that of
Mycoplasma genitalium, the smallest known free-living bacterium [
27]. The bacterium’s genome is quite small compared to the human genome, having 582,970 base pairs in 521 genes. Full genomes are difficult to synthesize because long strands of manufactured DNA tend to break. A new method was used to harness the DNA repair mechanism of yeast to stitch the full genome together. The bacteria’s DNA was also tagged with a watermark to indicate its source. In fact, there is ample room in synthesized DNA to include full documentation, licensing, liability, and other information. The sixty-four available codons could be mapped easily to the letters of the alphabet, numerals 0-9, and other symbols and characters. Some additional steps towards minimal genomes were achieved in September 2009 by cloning and modifying bacterial genomes in yeast [
28]. This was important as the first example of scientists being able to transfer genomes between branches of life, from a prokaryote to a eukaryote and back. The next step is to insert a synthesized genome into an empty nucleus and boot a cell to life.
3.2.4. Applications of synthetic biology
Some of the most important near-term applications of synthetic biology could be in energy and public health. There are several efforts to develop improved versions of fossil fuels which can be substituted easily into the existing global fuel infrastructure at approximately the same cost. Pilot plants are operating and commercial introduction is expected in 2011. Sapphire Energy and Synthetic Genomics are working with algal fuel, extending the highly efficient natural process of algae creating petroleum through photosynthesis. Amyris Biotechnologies is using synthetic biology to generate ethanol, and LS9 is synthesizing carbohydrates into petroleum with designer microbes. In the farther future, late-generation biofuels are contentious but already being envisioned by companies like Synthetic Genomics, employing carbon dioxide (CO2) as a feedstock for bacteria to convert into methane, using molecular hydrogen as the energy source. In public health, synthetic biology could be applied in the more rapid manufacture of biologics such as antibodies, proteins, and cells, and potentially used for the many eventual custom therapies of personalized medicine.
3.3. Whole cell characterization
DNA sequencing and synthesizing are near-term applications but ultimately, the goal is to see every aspect of living cells
in vivo on-demand. Work is beginning on these next steps, to examine at minimum the methylome (what portions of DNA are turned on and off in certain ways), the transcriptome (the RNA complement of DNA that has been transcribed), the proteome (what genetic messages have been expressed and translated into proteins), and the metabolome (byproducts of cellular activities). Some key progress has been made in mapping the full methylome of two human cell types [
29] and in the targeted analysis of specific proteins using mass spectrometry rather than traditional shotgun approaches [
30]. New approaches and tools are critical to advance. An exceptional development has occurred in electron microscopy that may allow the noninvasive molecular-resolution imaging of live samples (
Figure 6) [
31]. Usually electron microscopy is a destructive technique as the electron beam may destroy the sample in the process of inspecting it.
4. Biomedical applications
Following conceptual shifts in science and the fundamentals of working with DNA, biomedical applications is the third macroscopic phenomenon in the process of engineering life into technology. Here, the term biomedical applications includes the contemplation of the eventual understanding, management, and augmentation of all biological processes. While there are many advances towards this end, the four fields with the most pronounced and relevant progress are nanomedicine, surgical intervention, regenerative medicine, and aging. Once all biological processes are understood and manageable, there is no reason why human lifespan and healthspan (high-quality health years) could not be indefinite. Having more durable, reliable, longer-living, controllable, and augmentable biological bodies is a key step in engineering life into technology.
4.1. Nanomedicine
Many contemporary science fields are conducting work at the nanometer scale, including biomedicine. Nanomedicine is any health application in the 1-100 nanometer (nm) scale. To put this in perspective, a human hair is 80,000 nm wide, the limit of human vision is 10,000 nm, a virus is 50 nm wide, DNA is 2 nm wide, and an atom is 0.2 nm wide on average. Some of the most important areas in nanomedicine are described below in the order of their current degree of deployment: nanoparticles, quantum dots, DNA nanotechnology, biomolecular interface technologies, and medical nanorobotics.
4.1.1. Nanoparticles
Nanoparticles are emerging as an important mechanism for disease diagnosis and treatment, particularly in cancer, due to their cell-specific targeting capabilities (
Figure 7) [
32]. In cancer treatment, nanoparticles are made from a variety of materials including gold [
33], calcium phosphate, and carbon nanotubes [
34]. The nanoparticles may carry a payload and be coated on the outside with targeting molecules such as folic acid. The particles are injected into the body and within a few hours, locate and bind to the cancer cells. Cancer cells are relatively easy to target since they overexpress folic acid receptors. Once the nanoparticles attach to the cancer cells, they disgorge any payload through nanopores. Magnetic or other properties of the nanoparticles can then be engaged in conjunction with traditional radiation therapies to improve efficacy as disease cells are more precisely eradicated with minimal disruption to nearby healthy cells [
35]. In cancer diagnosis, nanoparticles may be more sensitive than traditional immunoassays for determining the presence of antigens in blood serum [
36]. This methodology could be extended to diagnose a variety of residual diseases such as HIV and mad cow disease.
4.1.2. Quantum dots
A second area of nanomedicine is quantum dots. Quantum dots are small structures which can contain electrical charge, essentially nanocrystals or mini-semiconductors, 2-10 nanometers (10-50 atoms) in diameter. They are so small that the addition or removal of an electron changes their properties in a useful way, such as by glowing when stimulated by an external source like ultraviolet (UV) light. The main application of quantum dots is in the lighting industry, but they can also be used in biomedicine as a potential successor to the organic dyes now used ubiquitously in medical research (
Figure 7). Organic dyes degrade quickly and have a limited spectrum of colors. Quantum dots stay in the body longer and allow fine structure viewing of cells; however have more biocompatibility issues than organic dyes.
4.1.3. DNA nanotechnology
A third interesting and foundational field of nanomedicine is DNA nanotechnology, using DNA as a structural building material rather than as a carrier of genetic information. Interlinking strands of DNA can be formed into standard building blocks like the Holliday junction (
Figure 7), which can then be used to fashion structures such as tiles, platforms, and lattices. One potential benefit of using DNA as a building material is that it allows construction in 3-D, a challenging nanoscale problem with few existing tools. Efforts are underway to harness the natural kinetics and chemical properties of DNA reactions for consistent design and construction [
37]. DNA nanotechnology could be used as an engineering material for nanoscale circuits, structures, and motors, and has a broad application area ranging from computing and basic materials to nanomedicine. There are several examples of the use of DNA nanotechnology for novel biomedical engineering purposes. First, a nanoscale DNA box was created that has a controllable lid which could be used as a more precise nanoparticle drug delivery system [
38]. A second recent innovation was the creation of a bipedal, autonomous DNA "walker" that can mimic a cell’s transportation system and could possibly walk up and down DNA to specific locations to facilitate repair or deliver cargo [
39]. A third advance was the building of nanoscale rotary motors which could possibly be used to generate ATP (
Figure 7) [
40].
4.1.4. Biomolecular interface technologies
Another fundamental development in nanomedicine is biomolecular interface technologies. Working at the molecular scale is allowing and indeed requiring the integration of organic and inorganic material, a core building block for engineering life into technology. The biomolecular interface focuses on areas such as the physical chemistry of cell surfaces, cell-substrate adhesion, and engineered biomimetic surfaces [
41]. Two interesting examples of advances in biomolecular interface technologies are genetically-engineered peptides and mechanically-interlocked structures such as catenanes and rotaxanes.
Genetically-engineered peptides (GEPIs) are able to join organic and inorganic material by regulating cell behavior at interfaces. This is accomplished by modifying the surface chemistry of the cell, for example, immobilizing bioactive molecules that cause infection. This allows improved binding and reduced infection in biomedical implants [
42]. Catenanes and rotaxanes are examples of mechanically-interlocked molecular structures which are built from two components, one organic, such as an amine, and one inorganic, such as a metal (
Figure 7). These organic-inorganic hybrids offer the ability to construct large carbon-based molecules that have the electronic, magnetic, and catalytic properties of inorganic matter and could be used useful in a variety of applications ranging from quantum computing to disease detection and treatment [
43].
4.1.5. Medical nanorobotics
A fifth interesting field of nanomedicine is medical nanorobotics. Significant advances may be accomplished with the reprogramming of natural biological processes, but to significantly enhance human capabilities, to really engineer life into technology, nanorobots (nanoscale machines) may be required. A variety of nanorobot species have been designed by Robert Freitas [
44,
45] which could eventually supplement or replace several biological functions (
Figure 8). Some of these include respirocytes as artificial red blood cells [
46], microbivores as artificial white blood cells or phagocytes [
47], clottocytes as artificial platelets [
48], vasculocytes and cleaners for artery repair, and chromallocytes as gene delivery vectors for chromosome replacement [
49]. Many innovations would be necessary to build nanorobots, particularly molecularly precise nanotechnology [
50] and nanofactories [
51]. One of the first steps is demonstrating positional assembly and mechanosynthesis, essentially the ability to add and remove atoms at will and produce atomically precise structures [
52].
In the longer term, nanorobots could facilitate the engineering of the human body into technology. The human body 2.0 could include the redesign of the digestive system. Nanorobots could travel in and out of the skin, interacting with the digestive system to cycle nutrients and waste. Blood-based nanorobots could supply the precise nutrients needed. Eating could become mainly recreational as fat insulin receptors and other cellular mechanisms would be engaged to control fat accumulation [
6]. The human body 3.0 could include the redesign of the circulatory system [
53]. The heart, lungs, and blood could become obsolete. Respirocyte nanorobots could deliver oxygen to the cells without requiring a liquid-based medium [
6]. At this stage, the upper esophagus, mouth, and brain would remain, together with the skin, muscle, skeleton, and their corresponding parts of the nervous system. The human body 4.0 could target the redesign of the nervous system including the brain.
4.2. Surgical intervention and regenerative medicine
While nanomedicine may be able to increasingly respond to pathologies from within the body, other interventions will continue to require the direct access of surgery for disease treatment and the installation of engineered tissues, organs, and other replacement parts. Two surgical areas have been demonstrating significant advance, robotic surgery and radiosurgery. Robotic surgery, using the da Vinci robotic surgery platform for example, is typically carried out by making small incisions in the body and inserting thin tubes with robotic arms. The physician then performs the surgery from a computer terminal, manipulating the robotic arms. Minimally-invasive robotic surgery is now used instead of open surgery about 80% of the time for certain procedures, however one study found more unwanted side effects with robotic surgery, suggesting that it may not yet be a fully superior approach [
54]. Another surgical platform, not even requiring physicians or other personnel in the treatment room is cyberknife radiosurgery. Physicians use imaging data and computational models to design a treatment plan ahead of time. In the treatment room, a small linear accelerator is used to generate radiation vectors to treat tumors [
55]. 3-D cameras adjust in real-time to patient breathing and other movements, allowing even deep neurological tumors to be treated with precision [
56].
As biological tissues and organs wear out and break down, regenerative medicine could be an important alternative for obtaining custom replacement parts that are less likely to become infected or rejected. Organ regeneration is a challenging problem. The first and most straightforward organs to grow are those that are hollow and do not require vascularization. Scientists have been able to generate human bladders, blood vessels, and heart valves in the lab [
57]. For vascularized organs, novel 3-D organ printing methodologies are under development using only biologics (cells and cell products). A scaffold-free tissue engineering process could be used to print 3-D tissues and organs which could be vascularized prior to implantation, relying on developmental biology to trigger the cells to fuse and self-assemble into organs [
58].
4.3. Aging
Managing daily health with preventive medicine and resolving acute and chronic disease issues as they arise with nanomedicine, surgical intervention, and regenerative medicine may significantly extend the ability to control human health, but the deeper longitudinal processes of aging likely require some additional specialized approaches.
4.3.1. Pathology of aging
Increasingly, aging is being understood as a systems biology problem involving diverse mechanisms. In younger organisms, homeostatic processes automatically manage problems as they arise, but in older organisms, the resolution processes do not work as well. When cells become damaged as a consequence of aging, they can either self-destruct through apoptosis (regulated cell death) or become senescent (living on without dividing). Senescent cells persist in tissues, where they may secrete inflammatory proteins. Many major age-related diseases, including atherosclerosis, heart attack, stroke and metabolic syndrome, share an inflammatory pathogenesis. The build-up of senescent cells can lead to both degenerative disease (aging) and hyper-proliferative disease (cancer) [
59,
60]. The SENS (Strategies for Engineered Negligible Senescence) Foundation has distilled the various pathologies of aging into seven concrete problems: DNA mutations in the cell nucleus and mitochondria, waste that builds up inside and outside cells, cell loss/atrophy, death-resistant cells, and extracellular crosslinks (cells that become stuck together) [
61].
4.3.2. Potential remedy of aging
A variety of forward-looking solutions have been suggested for the remedy of aging, ranging from biological therapies to nanomedical robotics. These ideas could be quite challenging to implement, particularly with current technical capabilities, and skepticism about their feasibility exists.
The SENS Foundation proposes biological therapies related to genes, enzymes, cells, and the immune system. Enzyme therapy and immune system vaccinations could be used to help the body rid senescent cells that escape immune system detection [
62], perhaps 10-15% of senescent cells. Enzyme therapies are also being investigated to address waste accumulation [
63]. Enzymes could be delivered to the sites of amyloidosis [
64], atherosclerosis [
65], and macular degeneration to bind with plaques and be expelled automatically through the body’s natural processes. Other therapies such as extracellular matrix turnover enhancement could be used to break built-up crosslink structures [
66].
More contentious solutions are proposed for the remaining problems of aging. Cell therapies could revive lost and atrophied cells, strengthen cells, and restore the elasticity of the extracellular matrix. Regenerative medicine could be used to make replacement tissues and organs. To combat DNA mutations in the mitochondria, gene therapies could be used to relocate the relevant mitochondrial genes (related to the generation of thirteen proteins) to the more-protected nucleic DNA for expression. DNA mutations in the cell nucleus that could potentially trigger cancer are an especially challenging problem. A radical proposal is that perhaps abnormal cell growth could be limited by deleting certain genes related to the ALT (alternative lengthening of telomeres) gene and telomerase enzyme expression. Periodic rejuvenation therapies (e.g., each five to ten years) could potentially reseed stem cells to ensure normal cell division [
61].
In nanomedicine, nanorobot-based cures for all major pathologies of aging have been proposed [
67]. Medical nanorobotics is ultimately envisioned be able to control all critical molecular events in the human body through targeted treatments to individual organs, tissues, cells, and intracellular components. Specific to human morbidity and aging, these mechanisms could be use to cure disease, reverse trauma, and repair cells. Senescence could be managed by removing waste, restoring cell capability, and repairing DNA damage.
5. Intelligence
Following the conceptual shifts in science, the fundamentals of working with DNA, and biomedical applications, intelligence is the fourth macroscopic phenomenon in the process of engineering life into technology. There are four key areas: neuroscience, artificial intelligence and robotics, whole brain simulation, and human-machine integration interfaces.
5.1. Neuroscience
The prevailing technique in behavioral and cognitive neuroscience is neuroimaging. Some contemporary themes in neuroimaging include spatiotemporal accuracy, pattern classification, imaging signals, and resting states [
68]. A useful resource is Wiley’s journal of Human Brain Mapping. Improvements in fMRI, CT, and PET scans have been extending the working knowledge of the brain through higher resolution and quicker image reconstruction. When destructive techniques are possible, the neuroimaging of tissue volumes may be conducted in an attempt to generate the connectome, a circuit diagram of the brain mapping each synapse and neural connection [
69]. Knife-edge scanning microscopy (KESM) has demonstrated synapse and vesicle detection in 300 nm wide brain tissue slices. Mapping stimulus to brain response and vice versa is another important area of current investigation [
70]. Applications range from pain self-management explored by companies like Omneuron [
71], to neuromarketing assessed by companies like EmSense, NeuroFocus, NeuroCo, and NeuroSense [
72].
5.2. Artificial intelligence and robotics
The second important area of intelligence research is building intelligence from scratch in software and robotic forms.
5.2.1. Artificial intelligence
Artificial intelligence (AI) is a branch of computer science dealing with the simulation of intelligent behavior in computers. AI research has many sub-areas such as natural language processing, information representation, pattern recognition, knowledge engineering, and machine learning.
Some of the main approaches to AI include expert systems, neural networks, Bayesian models, genetic algorithms, and hierarchical temporal memory. These approaches are all essentially different methods of absorbing information and processing it into actions. One of the areas with the highest level of current activity is genetic algorithms [
73]. Hierarchical temporal memory (HTM) is an interesting emerging approach with recent research framing the brain as a reward system as apposed to a memory system [
74]. A Silicon Valley-based company, Vitamin D (
http://www.vitamindinc.com), is implementing HTM technology for home and office video surveillance. Some of the biggest advances in artificial intelligence have been developed using non-traditional approaches. For example, Google has been making continuous progress in natural language machine learning by applying statistical methods to the large datasets that have arisen on the web [
75].
AI is used in many aspects of daily life, for example in credit card fraud detection, factory automation, and seismographic data examination. The Association for the Advancement of Artificial Intelligence (AAAI) tracks innovative AI applications as presented in conference papers, and lists 438 as of November 2009. Thirty-two were added in 2009, four deployed applications such as a tool for gas turbine maintenance scheduling, and twenty-eight emerging applications [
76].
AI may be categorized in two ways: strong, and weak or narrow. Most of the funding and application activity is in narrow AI, using intelligent systems to focus on specific problems. Strong AI, intelligent systems that can be used for general purpose problem solving, is a nascent field and some formalized theories of universal general intelligence are under development [
77]. There is debate about a variety of issues, including how moral or friendly AI could be created that would be beneficent to humans.
5.2.2. Robotics
Robotics is the technology dealing with the design, construction, and operation of robots in automation. Like artificial intelligence, there are a variety of subfields covering the various elements and their integration. Embedded systems, for example, is the field linking computational and physical components. Some of the most important current applications of robotics are in military use, factory automation, telepresence, entertainment, and human interaction. Some contemporary trends include bipedal robots, autonomous robotics, and swarm computing. Bipedal walking, instead of navigating around on wheeled or multi-legged bases, could open up a variety of new applications for robotics. Similarly, autonomous robots could execute tasks at a higher level of abstraction with less of a control burden, and self-organize activity across disparate computing nodes, for example in wireless sensor networks. Swarm computing could allow the efforts of multiple robots to be coordinated, for example in warehouse automation and RoboCup soccer.
The U.S. military’s current deployment of robots includes 7,000 unmanned aerial vehicles (UAVs) such as the Predator drone and 12,000 unmanned ground vehicles (UGVs) such as the PackBot [
78]. Boston Dynamics has developed several robots for military use. One is the BigDog, a quadruped robot that can walk, run, and climb on rough terrain and carry heavy loads [
79]. More recently, the company has developed the PETMAN bipedal robot (
Figure 9) which balances dynamically using a human-like walking motion. The PETMAN is to be used initially for testing chemical protection clothing by walking and climbing like a human. Another example of military robotics is the DARPA Grand Challenge, where there have been three rounds of competition for unmanned navigation vehicles, lastly in an urban environment [
80]. An important issue has arisen as to how ethics submodules may be successfully incorporated into autonomous robots and is reviewed extensively in a 2008 report commissioned by the U.S. Department of Navy’s Office of Naval Research [
81].
A second important area is scientific and industrial robotics, with a current focus on mobility. A robot scientist, Adam (
Figure 9) has been developed to design and carry out lab experiments. In mobile robotic solutions for warehouse automation, a leader is Kiva Systems, using robots to organize, manage, and move inventory (
Figure 9). The robots cooperate using swarm behavior by reading barcodes on the floor and using other messaging systems. There are additional examples of robots intended for use in corporate or health care environments. Willow Garage’s PR2 (Personal Robot 2) can execute tasks such as autonomously opening doors and locating and plugging itself in to power outlets (
Figure 9) [
82]. AnyBots offers a corporate telepresence robot and has a bipedal robot under development.
There is also a research focus on creating robots with emotional intelligence for human interaction. Two notable laboratories are MIT’s Personal Robots Group and Hanson Robotics. MIT has created robots such as Leonardo which has 50 independently controlled servo motors creating a full range of facial expressions. Hanson Robotics’ Albert-Hubo, Zeno, and other robots have life-like skin created from frubber, a nanoporous materials advance in elastic polymers (
Figure 9) [
83]. For consumer use, robots are starting as small appliances such as the Roomba and Neato for home vacuuming, the Rovio for home security, and toys such as the Furby, Aibo, and Kondo.
5.3. Whole brain simulation
One example of the expanded notion of science that includes statistical and computational models is the effort to understand and simulate the behavior of brains in software. The Whole Brain Emulation Roadmap attempts to enumerate the many detailed steps that may be required, together with a review of the challenges and potential progression necessary for the whole brain emulation of a human [
84]. Assessing the accuracy of software neural simulations is challenging and the focus of current research. For example, one project looks at the projectome, the complete set of white matter fascicles (bundles of nerve fibers), and builds a parallel algorithm for global projectome evaluation that takes into account problems such as global prediction error and volume conservation [
85]. The leading effort to create a software-based synthetic brain is the Blue Brain project at the Brain Mind Institute of the École Polytechnique Fédérale de Lausanne in Switzerland.
One of the first critical milestones in the Blue Brain project was achieved in 2007 by building an accurate computer-based replica of one neocortical column of a rat brain. The research team claims that the neocortical column (NCC) is the critical and modular component of the brain. The NCC has between 10,000 and 100,000 neurons depending on the species and neocortical region, and millions of NCCs comprise the full brain. Another milestone was reported in November 2008, that the project was able to emulate the full cortex of a two-month old rat at one-tenth real-time speed on a dedicated IBM Blue Gene L supercomputer [
86]. In July 2009, additional progress was reported regarding the fidelity and activity of the simulated rat brain, and the occurrence of some spontaneous behavior such as cells pulsing in unison [
87]. The simulated rat brain has 10,000 neurons per NCC with 10 million connections. By comparison, the human brain’s 100 billion neurons are estimated to have 100 trillion connections or synapses. The team estimates that it will take another ten years, until 2020, to emulate a full human brain at real-time speed.
Like many projects, Blue Brain is multidisciplinary and has been made possible through advances in related technologies. Neuroimaging, infrared differential interference control (infrared-DIC) videomicroscopy to record live synapse activity, and complex neuronal simulation software [
88] have been critical to the Blue Brain project. The underlying technologies are often Moore’s Law-based and are likely to continue to extend the capabilities of the project. For example, in October 2009, researchers at Delft University announced a camera sensor able to detect single photons and film action at one million frames per second. This camera could be used to capture the microsecond-long pulse-like nerve signals that travel through networks of neurons at up to 180 kilometers per hour, and possibly accelerate the live mapping activities of the Blue Brain and other neurosimulation projects [
89,
90].
5.4. Human-machine integration interfaces
The fourth important vector of intelligence research is integrating life and technology through human-machine interfaces like brain-computer interfaces (BCIs) and body area networks (BANs). A BCI is a direct communications pathway between the brain and an external device. BCIs are in use in two principal applications: invasive cures and noninvasive enhancements. First, neuroprostheses like the BrainGate neural interface from Cyberkinetics may be surgically implanted to allow severely motor-impaired individuals to communicate via thought. Signals in the motor cortex are read and translated into on-screen cursor movements or wheelchair commands [
91]. Second, noninvasive externally-worn BCIs such as the EEG-based BCI2000 may be used to generate virtual cursor movement [
92], or possibly allow thought-driven video game play [
93].
Subsequent generations of BCIs might be used for neuroengineering, enhancing brain function by establishing bi-directional communication and processing between the mind and external devices. Eventually, brain co-processors might be able to intelligently supplement the brain’s activity. Tackling new tiers of pathologies could be possible, addressing areas like stroke rehabilitation, addiction, and chronic pain, together with the general goal of enhancing cognitive function. Some of the issues in implementing two-way brain co-processors include a means of collecting data and identifying brain states, and mechanisms for beneficially perturbing cognitive function, for example with optical beams or magnetic fields [
94].
Body-area networks (BANs) are also envisioned as a key means of integrating life and technology, bringing processing and communications on-board the person for medical, consumer electronics, entertainment, and other applications. At present, BANs consist of one or a few wearable or implanted biosensors gathering basic biological data and transmitting it wirelessly to a computer [
95]. The IEEE’s BAN communication standards protocol is 802.15.6. Some medical examples of BANs include disposable medical BANs [
96] for measuring blood pH, glucose, oxygen levels, and temperature, and CardioMEMs, implantable wireless sensing devices less than one tenth the size of a dime for monitoring heart failure, aneurysms, and hypertension [
97]. A consumer application of BANs is health activity monitors such as the FitBit and DirectLife [
98], and to some extent smart phones. All three contain accelerometers that can measure movement and activity. The next phases of BANs could be enabled by continued electronics miniaturization and next-generation communications networks (WiMAX, 4G, and beyond). In the farther future, BANs could include larger more complex networks of intercommunicating sensors and eventually autonomous sensors with two-way broadcast.
Biocompatibility and bandwidth are important concerns for human-machine integration interfaces, particularly implanted interfaces. However, the biggest challenge is energy, providing adequate ongoing power to devices. Several interesting methods of power generation are being investigated including thermal, vibrational, radio frequency (RF), photovoltaic (PV), and bio-chemical energy. ATP could possibly provide power to implanted devices, for example using DNA nanotechnology to synthesize ATP with nanoscale rotary motors [
40], or nanodevices to produce ATP from naturally circulating glucose [
99].
6. Discussion of a complex dynamical system: engineering life into technology
Section 2,
Section 3,
Section 4 and
Section 5 provided a summary of the macroscopic and microscopic elements of contemporary science advances that are contributing to the engineering of life into technology. This section now discusses these research advances in the broader context of their involvement as elements of a complex dynamical system, how a complex dynamical system may be managed, and how a phase transition in intelligence might be characterized. A complex systems framework has been applied here for two reasons. First, the engineering of life into technology closely fits the definition and dynamics of a complex dynamical system as described below. Second, the techniques for managing complex dynamical systems could be well-suited to identifying and managing out of bounds conditions that could occur in the process of engineering life into technology.
6.1. Engineering life into technology is a complex dynamical system
Engineering life into technology meets the comprehensive definition of a complex system provided by complexity theorist Melanie Mitchell: “a system in which large networks of components with no central control and simple rules of operation give rise to complex collective behavior, sophisticated information processing, and adaptation via learning or evolution [
100].”
6.1.1. Complex collective behavior
Engineering life into technology is a system with large networks of components with no central control where simple rules of operation are giving rise to complex collective behavior. From the simple rules of human curiosity and the construct of trying to understand the world around and within us, at least three examples of complex collective behavior have occurred. First, the seemingly disparate networks of components exhibit the existence of a point attractor. All parts of the system can be construed as moving towards the same goal, the complete mastery of biology. The mastery of biology could make it possible to understand, manipulate, control, manufacture, and augment biology. Not just control over biology, but its explicit enhancement is likely required for a phase shift in intelligence.
Second, new system-wide patterns have emerged that are greater than the sum of the parts. Each microscopic element is advancing in its own domain, and may also serve as an enabler or required input for another network component. For example, DNA sequencing is an input to preventive medicine, nanomedicine is an input to neuroscience imaging, and biomolecular interface technologies are an input to nanomedicine and BCIs/BANs. The network elements are becoming increasingly interlinked. As advances occur in different areas, they could ripple through the system quickly and catalyze faster next-level responses from other groups of network elements. Third, the network elements are together redefining the notions of science, the scientific method, and how science is conducted, having an impact both within the complex dynamical system and more broadly on society.
6.1.2. Sophisticated information processing and signaling
Sophisticated information processing and signaling is taking place in the complex system of engineering life into technology. The influence of information processing is clear as many fields of science have become rigorously computational. Informatics techniques are used in research design, data collection, processing, analysis, and results communication. The large data sets and small size scales in many fields mean that progress would not be possible without information processing. New levels of both information processing and communications technologies will likely be required to extend current research. For example, in human genomics, first a few SNPs were genotyped, then one whole human genome was sequenced. Now SNP genotyping data exists for on the order of tens of thousands of people, and whole human genome sequences for only a handful of people. Next there could be a wider genotyping and eventual whole genome sequencing of millions of people, with both normal and disease samples (
i.e., cancer tumors). It will also be important to identify DNA methylations and structural variation, and gain additional tiers of visibility into the whole cell such as with the transcriptome, proteome, metabolome, and diseasome. The coming era of preventive medicine envisions collecting billions of biological data points on each human on a regular basis [
101].
Over time, science fields and operations could become increasingly information processing intensive. Computation could be embedded, making all matter smart and connected, including biological matter. Organic and inorganic matter could be joined at a variety of different levels. The current distinction between life and technology could become meaningless. Network elements have already been adapting to exponential changes in technology, and could become seamlessly integrated with technology and the more capacious processing power and quicker cycles of evolution it offers.
The influence of information signaling is also clear. The internet and social networking facilitate communication and collaboration, and also allow challenges and their potential pervasiveness to surface more quickly. For example, network elements, or science fields, have been highlighting to each other that some problems have become complex, combinatorial, and seemingly intractable. Traditional methods no longer work and new levels of discovery cannot be attained. Systems biology approaches have become a common theme at technical conferences. Some of the most notable fields with this challenge include cancer, aging, chronic disease, drug discovery, preventive medicine, neurological structure, and natural language processing. In addition, there are related problems of a general nature such as how so much complexity has arisen in nature from such small instruction sets. Information signaling may expediently surface problems and also identify potential solutions. Some proposed solutions to systems-level complexity problems in science include applying statistical methods to very large data sets [
75], evolving previously unknown equation sets [
102], and sampling the computational universe of all possible programs that could be human-usable [
10].
6.1.3. Adaptation via learning to improve survival
Adaptation via learning or evolution to improve the possibility of success or survival is critical in engineering life into technology and has been occurring in two ways: adaptation due to technological advance and adaptation due to the failure of traditional methods. First, keeping pace with advances in technology is the chief ongoing adaptation required by network elements. Exponential change in technology cycles has triggered not just quantitative differences but also qualitative differences. In the 1970s, a computer was the size of a room. Today, a mobile phone has the same processing power. It is hard to imagine that computers might not be microscopic and embedded in the body in the next several decades. Successful practitioners plan and execute projects with these dynamics in mind. Qualitative changes in technology are also requiring innovation in how science is conducted. Laboratory techniques used by today’s scientists may not resemble the methods of biology taught academically even a few years ago. For example, at present, most medical students are not trained in genomics, yet this is emerging as a key paradigm in medicine in the next few decades. The most successful individuals and fields are able to quickly identify and implement relevant external advances.
Second, network elements have had to evolve to survive because traditional methods of conducting science do not always yield results. Adaptations are occurring in the approaches and skills brought to bear on problems. Science fields have necessarily become more complex, and may simultaneously include reductionist, systems-level, and multidisciplinary approaches. The most successful labs may have a variety of skillsets including those from biology, chemistry, physics, materials, imaging, mathematics, statistics, computer science, and information technology.
6.2. Managing the complex dynamical system: engineering life into technology
Just as there are attempts to master and manipulate biology at every level of the complex dynamical system, so too could there be an attempt to manage the system as a whole. By definition, complex dynamical systems are not generally possible to control, but there may be some opportunities for management and guidance. At minimum, it could be possible to assess when the system is potentially heading or has gone out of bounds, and attempt to bring it back into alignment. A complex dynamical system and each of its constituent components may be in any of three patterns of behavior at any time (
Figure 10), according to the degree of certainty in the environment and agreement among agents.
Near the origin, where there is high certainty and agreement, the system is controlled and predictable, and traditional linear management methods may be used such as measuring progress towards a defined goal. At the other extreme, the top right corner, where there is low certainty and agreement, the system is random and unpredictable, and strong personal leadership and if necessary, force, may be the appropriate management methods. When the system is in the middle state, a complex dynamical system, a variety of management techniques may be employed, generally relying on the identifiable dynamics of the system. Two obvious management mechanisms could be identifying risks that threaten the existence of the system and its components, and outlining early warning signals of such conditions.
6.2.1. Global catastrophic risk management
The first and most important complex dynamical system management technique that could be applied is an overall management of existential risks, risks that pose a threat to human existence. There has been an effort to enumerate, quantify, and prioritize global catastrophic risks, some of which could arise in the process of engineering life into technology. Risks have been categorized into three areas: risks from nature, risks from unintended consequences, and risks from hostile acts [
104]. A global catastrophic risk conference survey asked participants to estimate the risk that at least one million people would die before the year 2100 from a variety of risks. Specific to the areas discussed in this analysis, participants estimated that there was a 10% risk of estimated death due to superintelligent artificial intelligence, 30% due to an engineered pandemic (
vs. 60% due to a natural pandemic), and 25% due to molecular nanotechnology weapons [
105]. One next step could be the development of quantitative web-based tools for monitoring global catastrophic risk, including crowdsourced information inputs, and forums for discussion regarding potential resolutions.
6.2.2. Early warning signals
The second complex dynamical system management technique that could be applied is identifying key indicators to serve as early warning signals for when different systems parameters may be heading out of bounds. Known dynamics of complex systems could be good key indicators and management levers, for example, sensitive dependence on initial conditions, tipping points, attractors, fractals, and agent-based behavior, some of which are pictured in
Figure 11. A model described for use in managing complexity in organizations has been applied here to the engineering of life into technology [
103].
Complex systems have sensitive dependence on initial conditions, sometimes known as the butterfly effect. Knowing the tight potential interlinkage of the loosely-coupled components of the system and how a small perturbation can trigger large impacts, there could be a mechanism for collecting information about millions of small events happening on the edges of the system. Automated algorithms could assess these events and positive occurrences (i.e., multidisciplinary breakthroughs, alternative approaches, documentation of approaches that do not work) could be encouraged through special or high-magnitude rewards. Other occurrences (i.e., incremental progress) could be de-emphasized or rewarded through alternative mechanisms.
Self-organized criticality, tipping points, is a related feature of complex dynamical systems. Tension may build up and eventually cause a shift in the entire system. Different kinds of incentives could be injected into the complex system to relieve or cause pressure in desired directions. For example, competition and innovation could be ramped up through high-profile prizes, grand challenges, and targeted funding.
Attractors are behaviors which evolve in complex systems over time, occurring on a point, periodic, strange, or random basis. Identifying these patterns may help to predict the impact of particular actions, and to target actions to either shift or enhance attractor patterns. For example, there could be an adverse public reaction to many of the advances in engineering life into technology. Spokespeople from a variety of backgrounds could be recruited and briefed in anticipation to provide calm and cogent communication outlining the benefits and risks of new technologies to different audiences.
Fractals are repeating behaviors at different scales, and could possibly be identified across the complex dynamical system. If certain patterns repeatedly lead to positive outcomes, perhaps they could be replicated in more locations and at different scales. For example, similar arguments may work for successful fundraising regardless of organization size or funding sources. Another example of deploying the concept of fractals is explaining new ideas with existing concepts (i.e., Facebook for the genome; upgrading the brain’s software; DIYgenomics).
Models of agent-based behavior may help to elicit the simple rules followed by individual agents that have resulted in beneficial collective behaviors (i.e., bees building hives or ants leaving pheromone trails). A simple rule might be noticed in the complex system, for example, that technology adoption was more positive and widespread when trained professionals delivered a new solution to individuals (i.e., genetic testing). Deploying simple rule sets across the complex system could produce behaviors that would contribute to the system as a whole. One example could be an alternative model of scientific publishing requiring that an unrelated team replicate any lab work before findings could be published.
6.3. The next node: characterizing a potential phase transition in intelligence
It is speculative but some inferences may be drawn as to the potential distinguishing features of a phase transition in intelligence. One suggestion is that there could be an intelligence explosion [
108], a vast expansion in the number and types of sentient beings. The current example of human intelligence may be one dot in a large possibility space of all potential intelligence. Other examples of intelligence may not resemble humans at all; analogous to the airplane being quite different from birds yet still able to fly. In the future, sentient beings could include a variety of augmented and unmodified humans, hybrids, and different machine species.
Another suggestion is that next-generation intelligence, superintelligence, may likely have greatly extended capabilities as compared with those of humans. Superintelligences may be able to think in three, four, or more dimensions, and could have more and better senses, for example, radar, ultrasound, and distributed, globe-spanning sensory systems. These senses might be able to identify abstract patterns in phenomena such as weather, energy use, demographics, and animal migration the same way a human can assess whether a commonly traveled road has more or less traffic than usual [
109]. One analogy is that as humans are to apes, superintelligences would be to current human-level intelligence. Superintelligences may think and learn a million times faster than humans, reading a book in ten seconds, or completing a human’s lifetime output in a few weeks, perhaps all the while self-improving through iterative cycles [
110].
7. Conclusions
Information optimization is a central dynamic in the universe, cycling between symmetry and imbalance to reach nodes of increasing complexity and diversity. Exponential advances in technology have given rise to a period of imbalance in contemporary information optimization as to the future of intelligence. In response to the broken symmetry, a complex dynamical system is developing, the engineering of life into technology. Many disparate network elements are coming together in this system. The network elements are increasingly interlinked and technology-integrated, with the common goal of biological mastery and augmentation, and could develop more order, balance, and symmetry to eventually self-organize into a phase transition in intelligence and information optimization.
The network elements include first, conceptual shifts in the notions of science, the scientific method, health, and preventive medicine; second, the fundamentals of the chemical software which operates life, DNA sequencing and synthesizing, and characterizing whole cell contents; third, the biomedical applications of optimizing biology through nanomedicine, surgical intervention, regenerative medicine, and longevity therapies; and fourth, extending intelligence through neuroscience, artificial intelligence and robotics, whole brain simulation, and human-machine integration interfaces.
The network elements exhibit the dynamics of a complex dynamical system: complex collective behavior, sophisticated information processing and signaling, and adaptation via learning and evolution to improve survival. The complex system may be managed, for example, by identifying risks that threaten the existence of the system and its components, and outlining early warning signals of such conditions. Specific attributes of the complex system could be used as management levers, for example manipulating sensitive dependence on initial conditions, tipping points, attractors, fractals, and agent-based behavior. Speculating on the nature of a phase transition in intelligence, the next node could feature superintelligences of many kinds with enhanced capabilities in a variety of dimensions ranging from sensory perception and processing ability to most importantly, rapid self-improvement.
Despite the robustness and resilience of the complex system of engineering life into technology, it may not be the ultimate source of a potential phase transition in intelligence. As progression towards a phase transition becomes more intense, different routes could become distinct. For example, there could be at least three approaches: engineering life into technology, simulated intelligence, and artificial intelligence. Right now, these approaches are intertwined and reciprocally enabling, but one could move ahead of the others. There are significant technical hurdles in executing simulated intelligence and artificial intelligence, but engineering life into technology could be quite challenging too, both the technical issues and the ethical, legal, and social issues (ELSI). The engineering of life into technology will need to proceed expediently to keep pace with technological advance, and to be able to contribute positively to a potential phase transition in intelligence.