Characterization of RNA-Like Oligomers from Lipid-Assisted Nonenzymatic Synthesis: Implications for Origin of Informational Molecules on Early Earth
Abstract
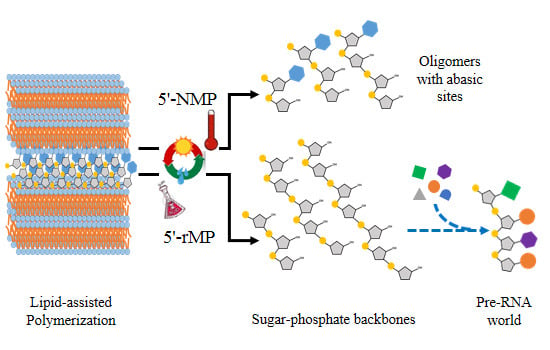
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Methods
2.2.1. Simulating Prebiotic Conditions
2.2.2. HPLC Analysis
2.2.3. Mass Analysis
3. Results
3.1. HPLC Analysis of Products

3.2. Effect of pH and Rehydrating Agent on Polymerization


3.3. Effect of Temperature on Polymerization

3.4. Optimum Dehydration Time for Polymerization

3.5. Optimum Reactant Ratios for Polymerization

3.6. Mass Analysis of Oligomers

| Peak No. | Assigned Species | Expected Mass | Observed Mass |
|---|---|---|---|
| 1 | Adenine | 136.0617 | 136.0629 |
| 2 | AMP | 348.0703 | 348.0701 |
| 3 | AMP + Na | 370.0523 | 370.0540 |
| 4 | Cyclic AMP Dimer minus (−) Base | 542.0683 | 542.0687 |
| 5 | AMP Dimer − Base | 560.0789 | 560.0791 |
| 6 | AMP Dimer − Base + Na | 582.0608 | 582.0610 |
| 7 | (AMP)2 | 695.1334 | 695.1333 |
| 8 | (AMP)2 + Na | 717.1153 | 717.1183 |
| 9 | AMP Trimer − 2 Bases | 772.0875 | 772.0829 |
| 10 | (AMP)2 + Dimer − base | 907.1420 | 907.1371 |
| 11 | Tetramer − 3 Bases | 984.0961 | 984.0882 |
3.7. Reaction with Other Nucleoside 5'-Monophosphates (5'-NMPs)
3.8. Reaction with Ribose 5'-Monophosphate (5'-rMP) to Form Sugar-Phosphate Oligomers


| Chemical Species | Expected Mass | Observed Mass |
|---|---|---|
| Cyclic rMP Dimer | 425.0244 | 425.0251 |
| Linear rMP Dimer | 443.0350 | 443.0363 |
| Cyclic rMP Trimer | 637.0330 | 637.0375 |
| Linear rMP Trimer | 655.0436 | 655.0424 |
| Cyclic rMP Tetramer | 849.0416 | 849.0469 |
| Linear rMP Tetramer | 867.0522 | 867.0532 |
| Cyclic rMP Pentamer | 1061.0502 | 1061.0449 |
| Linear rMP Pentamer | 1079.0608 | 1079.0615 |
| Cyclic rMP Hexamer | 1273.0588 | 1273.0591 |
| Linear rMP Hexamer | 1291.0694 | 1291.0681 |
| Linear rMP Heptamer | 1503.0780 | 1503.0774 |
4. Discussion
Acknowledgments
Author Contributions
Supplementary Materials
Conflicts of Interest
References
- Orgel, L.E. Prebiotic chemistry and the origin of the RNA World. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 99–123. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P. Mineral catalysis and prebiotic synthesis: Montmorillonite-Catalyzed formation of RNA. Elements 2005, 1, 145–149. [Google Scholar] [CrossRef]
- Menor-Salván, C.; Marín-Yaseli, M.R. Prebiotic chemistry in eutectic solutions at the water-ice matrix. Chem. Soc. Rev. 2012, 41, 5404–5415. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.D. Ribonucleotides. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Powner, M.W.; Sutherland, J.D.; Szostak, J.W. Chemoselective multicomponent one-pot assembly of purine precursors in water. J. Am. Chem. Soc. 2010, 132, 16677–16688. [Google Scholar] [CrossRef]
- Deamer, D.; Singaram, S.; Rajamani, S.; Kompanichenko, V.; Guggenheim, S. Self-assembly processes in the prebiotic environment. Philos. Trans. R. Soc. B 2006, 361, 1809–1818. [Google Scholar] [CrossRef]
- Engelhart, A.E.; Hud, N.V. Primitive Genetic Polymers. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Lahav, N.; White, D.; Chang, S. Peptide formation in the pre-biotic Earth: Thermal condensation of glycine in fluctuating clay environments. Science 1978, 201, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.B. Free energies and equilibria of peptide bond hydrolysis and formation. Biopolymers 1998, 45, 351–353. [Google Scholar] [CrossRef]
- Weber, A.L. Thermal synthesis and hydrolysis of polyglyceric acid. Orig. Life Evol. Biosph. 1989, 19, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Harshe, Y.M.; Storti, G.; Morbidelli, M.; Gelosa, S.; Moscatelli, D. Polycondensation kinetics of lactic acid. Macromol. React. Eng. 2007, 1, 611–621. [Google Scholar]
- Mamajanov, I.; MacDonald, P.J.; Ying, J.; Duncanson, D.M.; Dowdy, G.R.; Walker, C.A.; Engelhart, A.E.; Fernández, F.M.; Grover, M.A.; Hud, N.V.; et al. Ester formation and hydrolysis during wet-dry cycles: Generation of far-from-equilibrium polymers in a model prebiotic reaction. Macromolecules 2014, 47, 1334–1343. [Google Scholar] [CrossRef]
- Rajamani, S.; Vlassov, A.; Benner, S.; Coombs, A.; Olasagasti, F.; Deamer, D. Lipid-assisted synthesis of RNA-like polymers from mononucleotides. Orig. Life Evol. Biosph. 2008, 38, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Toppozini, L.; Dies, H.; Deamer, D.; Rheinstädter, M.C. Adenosine monophosphate forms ordered arrays in multilamellar lipid matrices: Insights into assembly of nucleic acid for primitive life. PLoS One 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Olasagasti, F.; Kim, H.J.; Pourmand, N.; Deamer, D.W. Non-enzymatic transfer of sequence information under plausible prebiotic conditions. Biochimie 2011, 93, 556–561. [Google Scholar] [CrossRef] [PubMed]
- DeGuzman, V.; Vercoutere, W.; Shenasa, H.; Deamer, D. Generation of oligonucleotides under hydrothermal conditions by non-enzymatic polymerization. J. Mol. Evol. 2014, 78, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.C.; Aldersley, M.F.; Delano, J.W.; Ferris, J.P. Mechanism of montmorillonite catalysis in the formation of RNA oligomers. J. Am. Chem. Soc. 2009, 131, 13369–13374. [Google Scholar] [CrossRef] [PubMed]
- Schrum, J.P.; Ricardo, A.; Krishnamurthy, M.; Blain, J.C.; Szostak, J.W. Efficient and rapid template-directed nucleic acid copying using 2′-amino-2′, 3′-dideoxyribonucleoside-5′-phosphorimidazolide monomers. J. Am. Chem. Soc. 2009, 131, 14560–14570. [Google Scholar] [CrossRef] [PubMed]
- Jordan, F.; Niv, H. Glycosyl conformational and inductive effects on the acid catalysed hydrolysis of purine nucleosides. Nucleic Acids Res. 1977, 4, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Zoltewicz, J.A.; Clark, D.F.; Sharpless, T.W.; Grahe, G. Kinetics and mechanism of the acid-catalyzed hydrolysis of some purine nucleosides. J. Am. Chem. Soc. 1970, 92, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, E.D.; Engelhart, A.E.; Chen, M.C.; Quarles, K.A.; Smith, M.W.; Lynn, D.G.; Hud, N.V. Intercalation as a means to suppress cyclization and promote polymerization of base-pairing oligonucleotides in a prebiotic world. Proc. Natl. Acad. Sci. USA 2010, 107, 5288–5293. [Google Scholar] [CrossRef] [PubMed]
- Vauthey, S.; Santoso, S.; Hwang, W.; Hartman, H.; Zhang, S. Self-assembly of Surfactant-like Peptides with Variable Glycine Tails to Form Nanotubes and Nanovesicles. Nano Lett. 2002, 2, 687–691. [Google Scholar] [CrossRef]
- Jakschitz, T.A.E.; Rode, B.M. Chemical evolution from simple inorganic compounds to chiral peptides. Chem. Soc. Rev. 2012, 41, 5484–5489. [Google Scholar] [CrossRef] [PubMed]
- Leman, L.; Orgel, L.; Ghadiri, M.R. Carbonyl sulfide-mediated prebiotic formation of peptides. Science 2004, 306, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Huber, C.; Wächtershäuser, G. Peptides by Activation of Amino Acids with CO on (Ni,Fe)S Surfaces: Implications for the Origin of Life. Science 1998, 281, 670–672. [Google Scholar] [CrossRef] [PubMed]
- Hitz, T.; Luisi, P.L. Liposome-assisted selective polycondensation of alpha-amino acids and peptides. Biopolymers 2000, 55, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Zepik, H.H.; Rajamani, S.; Maurel, M.C.; Deamer, D. Oligomerization of thioglutamic acid: Encapsulated reactions and lipid catalysis. Orig. Life Evol. Biosph. 2007, 37, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Yakhnin, A.V. A model for the origin of life through rearrangements among prebiotic phosphodiester polymers. Orig. Life. Evol. Biosph. 2013, 43, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Hud, N.V.; Cafferty, B.J.; Krishnamurthy, R.; Williams, L.D. The origin of RNA and “My Grandfather’s Axe”. Chem. Biol. 2013, 20, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Fuller, W.D.; Sanchez, R.A.; Orgel, L.E. Studies in prebiotic synthesis VII: Solid-state synthesis of purine nucleosides. J. Mol. Evol. 1972, 1, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Bean, H.D.; Sheng, Y.; Collins, J.P.; Anet, F.A.L.; Leszczynski, J.; Hud, N.V. Formation of a β-pyrimidine nucleoside by a free pyrimidine base and ribose in a plausible prebiotic reaction. J. Am. Chem. Soc. 2007, 129, 9556–9557. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mungi, C.V.; Rajamani, S. Characterization of RNA-Like Oligomers from Lipid-Assisted Nonenzymatic Synthesis: Implications for Origin of Informational Molecules on Early Earth. Life 2015, 5, 65-84. https://0-doi-org.brum.beds.ac.uk/10.3390/life5010065
Mungi CV, Rajamani S. Characterization of RNA-Like Oligomers from Lipid-Assisted Nonenzymatic Synthesis: Implications for Origin of Informational Molecules on Early Earth. Life. 2015; 5(1):65-84. https://0-doi-org.brum.beds.ac.uk/10.3390/life5010065
Chicago/Turabian StyleMungi, Chaitanya V., and Sudha Rajamani. 2015. "Characterization of RNA-Like Oligomers from Lipid-Assisted Nonenzymatic Synthesis: Implications for Origin of Informational Molecules on Early Earth" Life 5, no. 1: 65-84. https://0-doi-org.brum.beds.ac.uk/10.3390/life5010065