Whole-Genome Sequencing Analysis of a stx-Negative Escherichia coli O63:H6 Isolate Associated with Hemolytic Uremic Syndrome
Abstract
:1. Introduction
2. Materials and Methods
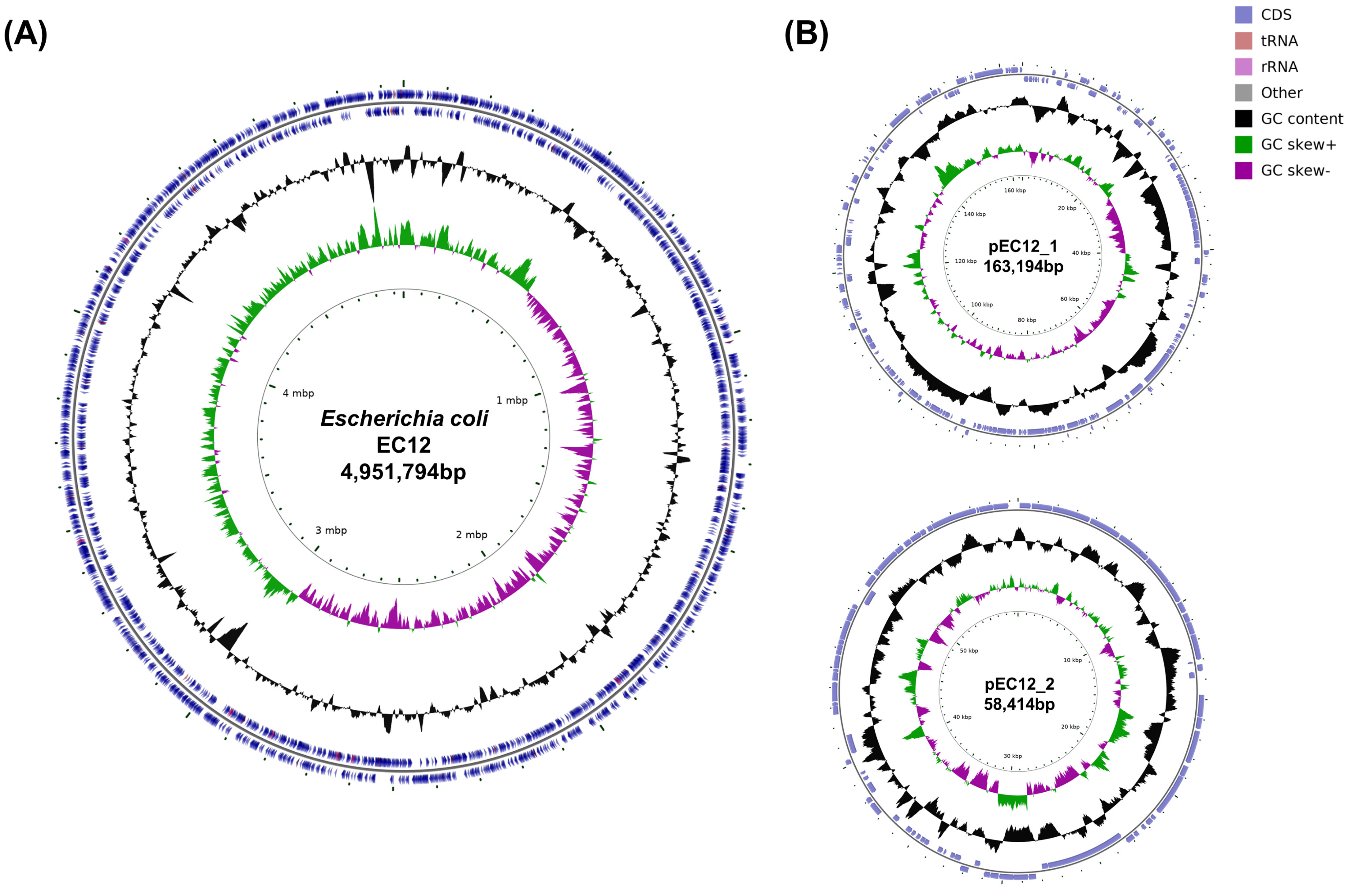
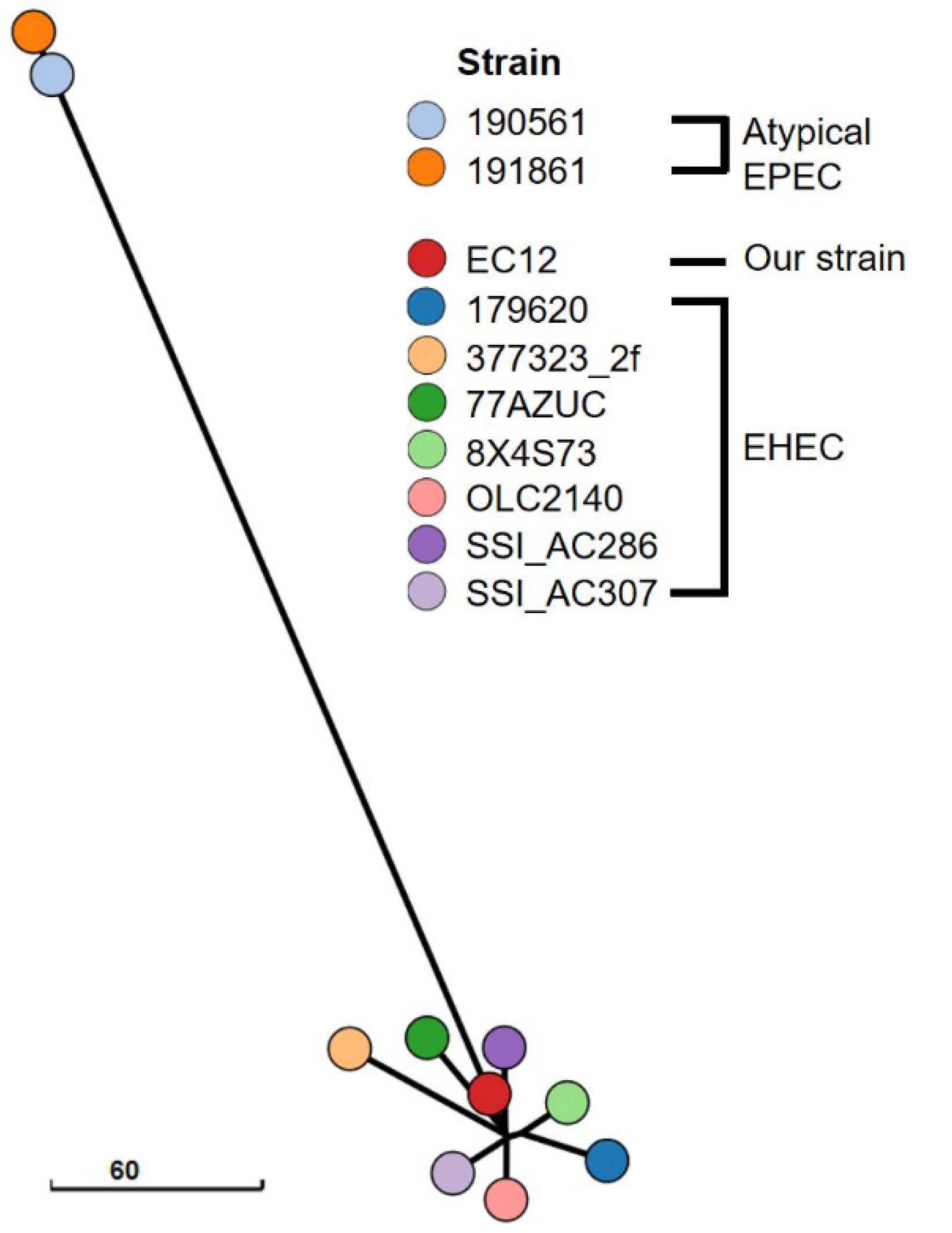
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Valilis, E.; Ramsey, A.; Sidiq, S.; DuPont, H.L. Non-O157 Shiga toxin-producing Escherichia coli—A poorly appreciated enteric pathogen: Systematic review. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2018, 76, 82–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, J.T.; Sowers, E.G.; Wells, J.G.; Greene, K.D.; Griffin, P.M.; Hoekstra, R.M.; Strockbine, N.A. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J. Infect. Dis. 2005, 192, 1422–1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prager, R.; Fruth, A.; Siewert, U.; Strutz, U.; Tschäpe, H. Escherichia coli encoding Shiga toxin 2f as an emerging human pathogen. Int. J. Med. Microbiol. IJMM 2009, 299, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Buvens, G.; De Gheldre, Y.; Dediste, A.; de Moreau, A.I.; Mascart, G.; Simon, A.; Allemeersch, D.; Scheutz, F.; Lauwers, S.; Piérard, D. Incidence and virulence determinants of verocytotoxin-producing Escherichia coli infections in the Brussels-Capital Region, Belgium, in 2008–2010. J. Clin. Microbiol. 2012, 50, 1336–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friesema, I.; van der Zwaluw, K.; Schuurman, T.; Kooistra-Smid, M.; Franz, E.; van Duynhoven, Y.; van Pelt, W. Emergence of Escherichia coli encoding Shiga toxin 2f in human Shiga toxin-producing E. coli (STEC) infections in the Netherlands, January 2008 to December 2011. Eurosurveillance 2014, 19, 26–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rauw, K.; Jacobs, S.; Piérard, D. Twenty-seven years of screening for Shiga toxin-producing Escherichia coli in a university hospital. Brussels, Belgium, 1987–2014. PLoS ONE 2018, 13, e0199968. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Hartland, E.L.; Leong, J.M. Enteropathogenic and enterohemorrhagic E. coli: Ecology, pathogenesis, and evolution. Front. Cell. Infect. Microbiol. 2013, 3, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochoa, T.J.; Contreras, C.A. Enteropathogenic E. coli infection (EPEC) in children. Curr. Opin. Infect. Dis. 2011, 24, 478–483. [Google Scholar] [CrossRef] [Green Version]
- Bielaszewska, M.; Köck, R.; Friedrich, A.W.; von Eiff, C.; Zimmerhackl, L.B.; Karch, H.; Mellmann, A. Shiga toxin-mediated hemolytic uremic syndrome: Time to change the diagnostic paradigm? PLoS ONE 2007, 2, e1024. [Google Scholar] [CrossRef]
- Ferdous, M.; Zhou, K.; Mellmann, A.; Morabito, S.; Croughs, P.D.; de Boer, R.F.; Kooistra-Smid, A.M.; Rossen, J.W.; Friedrich, A.W. Is Shiga toxin-negative Escherichia coli O157:H7 enteropathogenic or enterohemorrhagic Escherichia coli? Comprehensive molecular analysis using whole-genome sequencing. J. Clin. Microbiol. 2015, 53, 3530–3538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senthakumaran, T.; Brandal, L.T.; Lindstedt, B.A.; Jørgensen, S.B.; Charnock, C.; Tunsjø, H.S. Implications of stx loss for clinical diagnostics of Shiga toxin-producing Escherichia coli. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2018, 37, 2361–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [Green Version]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [Green Version]
- Friesema, I.H.; Keijzer-Veen, M.G.; Koppejan, M.; Schipper, H.S.; van Griethuysen, A.J.; Heck, M.E.; van Pelt, W. Hemolytic uremic syndrome associated with Escherichia coli O8:H19 and Shiga toxin 2f gene. Emerg. Infect. Dis. 2015, 21, 168–169. [Google Scholar] [CrossRef] [PubMed]
- van Hoek, A.; van Veldhuizen, J.N.J.; Friesema, I.; Coipan, C.; Rossen, J.W.A.; Bergval, I.L.; Franz, E. Comparative genomics reveals a lack of evidence for pigeons as a main source of stx2f-carrying Escherichia coli causing disease in humans and the common existence of hybrid Shiga toxin-producing and enteropathogenic E. coli pathotypes. BMC Genom. 2019, 20, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Strain | astA | bfpA | chuA | cif | eae | espA | espC | espF | espJ | gad | ibeA | nleB | nleC | ompT | stx | tccP | terC | tir | yfcV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC12 | + | + | + | + | + | + | + | + | + | + | + | − | + | + | − | + | + | + | + |
| 377323_2f | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + * | + | + | + | + |
| 179620 | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + * | − | + | + | + |
| 77AZUC | + | + | + | + | + | + | + | + | + | − | + | − | + | + | + * | − | + | + | + |
| 8X4S73 | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + * | + | + | + | + |
| SSI_AC286 | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + * | − | + | + | + |
| SSI_AC307 | + | + | + | + | + | + | + | + | + | − | + | − | + | + | + * | − | + | + | + |
| OLC2140 | − | − | + | + | + | + | + | + | + | − | + | − | + | + | + * | + | + | + | + |
| 190561 | − | − | + | + | + | + | + | + | + | + | + | + | − | + | − | − | + | + | + |
| 191861 | − | − | + | + | + | + | + | + | + | + | + | + | − | + | − | − | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.Y.; La, T.-M.; Kim, T.; Yun, S.A.; Lee, S.-W.; Huh, H.J.; Lee, N.Y. Whole-Genome Sequencing Analysis of a stx-Negative Escherichia coli O63:H6 Isolate Associated with Hemolytic Uremic Syndrome. Diagnostics 2021, 11, 1823. https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics11101823
Kim TY, La T-M, Kim T, Yun SA, Lee S-W, Huh HJ, Lee NY. Whole-Genome Sequencing Analysis of a stx-Negative Escherichia coli O63:H6 Isolate Associated with Hemolytic Uremic Syndrome. Diagnostics. 2021; 11(10):1823. https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics11101823
Chicago/Turabian StyleKim, Tae Yeul, Tae-Min La, Taesoo Kim, Sun Ae Yun, Sang-Won Lee, Hee Jae Huh, and Nam Yong Lee. 2021. "Whole-Genome Sequencing Analysis of a stx-Negative Escherichia coli O63:H6 Isolate Associated with Hemolytic Uremic Syndrome" Diagnostics 11, no. 10: 1823. https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics11101823






