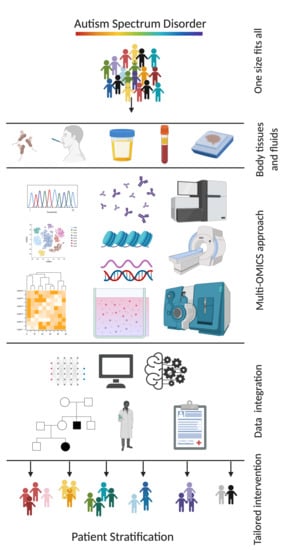
Paving the Way toward Personalized Medicine: Current Advances and Challenges in Multi-OMICS Approach in Autism Spectrum Disorder for Biomarkers Discovery and Patient Stratification
Abstract
:1. Introduction
2. Genetics of ASD: Understanding the Etiology and the Heritability
3. Non-Coding RNA’s as Biomarkers for ASD
4. Evidence of Epigenetics Modifications in ASD
5. Proteomics: A Fundamental Tool That Helps in Biomarker Discovery
6. Autoantibodies Biomarkers Suggest a Potential Molecular Sub-Class of ASD
7. Metabolomics and Gut Microbiome’s Biomarkers in ASD
8. Mitochondria Dysfunction in ASD
9. Brain Imaging, ERPs, and Eye-Tracking
10. Lessons from Other Diseases in Precision Medicine
11. Contemporary Challenges and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADHD | Attention deficit hyperactivity disorder |
| ADOS | Autism Diagnostic Observation Schedule |
| AGE | Glycation end products |
| ASD | Autism spectrum disorder |
| BA10 | Brodmann area 10 |
| CARS | Childhood Autism Rating Scale |
| CGH | Comprehensive genomic hybridization |
| CNV | Copy number variant |
| CSF | Cerebrospinal fluid |
| CVD | Cardiovascular diseases |
| DD | Developmental delay |
| ERP | Event-related potentials |
| GI | Gastrointestinal |
| T2DM | Diabetes mellitus type 2 |
| DSM-5 | Diagnostic and statistical manual of mental disorders- 5 |
| DZ | Dizygotic |
| fcMRI | Functional connectivity magnetic resonance imaging |
| FXS | Fragile X syndrome |
| GC | Gas chromatography |
| GWAS | Genome wide association studies |
| iASD | Idiopathic Autism spectrum disorder |
| ICD-10 | International Classification of Disease 10 |
| LC | Liquid chromatography |
| MALDI-TOF | Matrix-Assisted Laser Desorption Ionization Time-of-Flight |
| miRNA | microRNA |
| MS | Mass spectrometry |
| mtDNA | mitochondrial DNA |
| MZ | Monozygotic |
| ncRNA | non-coding RNA |
| NGS | Next generation sequencing |
| NDR | Neurodevelopmental regression |
| PDD-NOS | Pervasive developmental disorder not otherwise specified |
| PTM | Post-translational modifications |
| rRNA | Ribosomal RNA |
| RT-PCR | Reverse transcriptase-polymerase chain reaction |
| snoRNA | Small nucleolar RNA |
| snRNA | Small nuclear RNA |
| SNV | Single nucleotide variant |
| SV | Structural variants |
| TD | Typically developed |
| TLDA | TaqMan low-density array |
| TSC | Tuberous sclerosis complex |
| WES | Whole-exome sequencing |
| WGS | Whole-genome sequencing |
References
- Sharma, S.R.; Gonda, X.; Tarazi, F.I. Autism Spectrum Disorder: Classification, diagnosis and therapy. J. Pharmacol. Ther. 2018, 190, 91–104. [Google Scholar] [CrossRef]
- Hodges, H.; Fealko, C.; Soares, N. Autism spectrum disorder: Definition, epidemiology, causes, and clinical evaluation. Transl. Pediatr. 2020, 9, S55–S65. [Google Scholar] [CrossRef] [PubMed]
- Mandell, D.S.; Novak, M.M.; Zubritsky, C.D. Factors Associated with Age of Diagnosis among Children with Autism Spectrum Disorders. Pediatrics 2005, 116, 1480–1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrdlicka, M.; Vacova, M.; Oslejskova, H.; Gondžová, V.; Vadlejchova, I.; Kocourkova, J.; Koutek, J.; Dudova, I. Age at diagnosis of autism spectrum disorders: Is there an association with socioeconomic status and family self-education about autism? Neuropsychiatr. Dis. Treat. 2016, 12, 1639–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidovitch, M.; Levit-Binnun, N.; Golan, D.; Manning-Courtney, P. Late Diagnosis of Autism Spectrum Disorder after Initial Negative Assessment by a Multidisciplinary Team. J. Dev. Behav. Pediatr. 2015, 36, 227–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fusar-Poli, L.; Brondino, N.; Rocchetti, M.; Panisi, C.; Provenzani, U.; Damiani, S.; Politi, P. Diagnosing ASD in Adults without ID: Accuracy of the ADOS-2 and the ADI-R. J. Autism Dev. Disord. 2017, 47, 3370–3379. [Google Scholar] [CrossRef]
- Brondino, N.; Fusar-Poli, L.; Miceli, E.; Di Stefano, M.; Damiani, S.; Rocchetti, M.; Politi, P. Prevalence of Medical Comorbidities in Adults with Autism Spectrum Disorder. J. Gen. Intern. Med. 2019, 34, 1992–1994. [Google Scholar] [CrossRef]
- Warren, Z.; McPheeters, M.L.; Sathe, N.; Foss-Feig, J.H.; Glasser, A.; Veenstra-VanderWeele, J. A systematic review of early intensive intervention for autism spectrum disorders. Pediatrics 2011, 127, e1303–e1311. [Google Scholar] [CrossRef]
- Rossignol, D.A.; Frye, R.E. A review of research trends in physiological abnormalities in autism spectrum disorders: Immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and environmental toxicant exposures. Mol. Psychiatry 2012, 17, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Muhle, R.; Trentacoste, S.V.; Rapin, I. The Genetics of Autism. Pediatrics 2004, 113, e472–e486. [Google Scholar] [CrossRef] [Green Version]
- Smalley, S.L. Autism and tuberous sclerosis. J. Autism Dev. Disord. 1998, 28, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Bailey, D.B., Jr.; Raspa, M.; Olmsted, M.; Holiday, D.B. Co-occurring conditions associated with FMR1 gene variations: Findings from a national parent survey. Am. J. Med. Genet. A 2008, 146a, 2060–2069. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, G.B.; Lutz, R.E. Diagnostic yield in the clinical genetic evaluation of autism spectrum disorders. Genet. Med. 2006, 8, 549–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman, G.E.; Henninger, N.; Ratliff-Schaub, K.; Pastore, M.; Fitzgerald, S.; McBride, K.L. Genetic testing in autism: How much is enough? Genet. Med. 2007, 9, 268–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdul-Rahman, O.A.; Hudgins, L. The diagnostic utility of a genetics evaluation in children with pervasive developmental disorders. Genet. Med. 2006, 8, 50–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devlin, B.; Scherer, S.W. Genetic architecture in autism spectrum disorder. Curr. Opin. Genet. Dev. 2012, 22, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Acuna-Hidalgo, R.; Veltman, J.A.; Hoischen, A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 2016, 17, 241. [Google Scholar] [CrossRef] [Green Version]
- Persico, A.M.; Napolioni, V. Autism genetics. Behav. Brain Res. 2013, 251, 95–112. [Google Scholar] [CrossRef]
- Glessner, J.T.; Wang, K.; Cai, G.; Korvatska, O.; Kim, C.E.; Wood, S.; Zhang, H.; Estes, A.; Brune, C.W.; Bradfield, J.P.; et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009, 459, 569–573. [Google Scholar] [CrossRef]
- Roohi, J.; Montagna, C.; Tegay, D.H.; Palmer, L.E.; DeVincent, C.; Pomeroy, J.C.; Christian, S.L.; Nowak, N.; Hatchwell, E. Disruption of contactin 4 in three subjects with autism spectrum disorder. J. Med. Genet. 2008, 46, 176–182. [Google Scholar] [CrossRef] [Green Version]
- Farzin, F.; Perry, H.; Hessl, D.; Loesch, D.; Cohen, J.; Bacalman, S.; Gane, L.; Tassone, F.; Hagerman, P.; Hagerman, R. Autism Spectrum Disorders and Attention-Deficit/Hyperactivity Disorder in Boys with the Fragile X Premutation. J. Dev. Behav. Pediatr. 2006, 27 (Suppl. 2), S137–S144. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onay, H.; Kacamak, D.; Kavasoglu, A.N.; Akgün, B.; Yalcinli, M.; Kose, S.; Ozbaran, B. Mutation analysis of the NRXN1 gene in autism spectrum disorders. Balk. J. Med. Genet. 2016, 19, 17–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durand, C.M.; Betancur, C.; Boeckers, T.M.; Bockmann, J.; Chaste, P.; Fauchereau, F.; Nygren, G.; Rastam, M.; Gillberg, I.C.; Anckarsäter, H.; et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2006, 39, 25–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vignoli, A.; La Briola, F.; Peron, A.; Turner, K.; Vannicola, C.; Saccani, M.; Magnaghi, E.; Scornavacca, G.F.; Canevini, M.P. Autism spectrum disorder in tuberous sclerosis complex: Searching for risk markers. Orphanet J. Rare Dis. 2015, 10, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [Green Version]
- Vasu, M.M.; Anitha, A.; Thanseem, I.; Suzuki, K.; Yamada, K.; Takahashi, T.; Wakuda, T.; Iwata, K.; Tsujii, M.; Sugiyama, T.; et al. Serum microRNA profiles in children with autism. Mol. Autism 2014, 5, 40. [Google Scholar] [CrossRef] [Green Version]
- Cirnigliaro, M.; Barbagallo, C.; Gulisano, M.; Domini, C.N.; Barone, R.; Barbagallo, D.; Ragusa, M.; Di Pietro, C.; Rizzo, R.; Purrello, M. Expression and Regulatory Network Analysis of miR-140-3p, a New Potential Serum Biomarker for Autism Spectrum Disorder. Front. Mol. Neurosci. 2017, 10, 250. [Google Scholar] [CrossRef] [Green Version]
- Hicks, S.D.; Ignacio, C.; Gentile, K.; Middleton, F.A. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016, 16, 52. [Google Scholar] [CrossRef] [Green Version]
- Abu-Elneel, K.; Liu, T.; Gazzaniga, F.S.; Nishimura, Y.; Wall, D.P.; Geschwind, D.H.; Lao, K.; Kosik, K.S. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics 2008, 9, 153–161. [Google Scholar] [CrossRef]
- Hicks, S.D.; Carpenter, R.L.; Wagner, K.E.; Pauley, R.; Barros, M.; Tierney-Aves, C.; Barns, S.; Greene, C.D.; Middleton, F.A. Saliva MicroRNA Differentiates Children with Autism From Peers with Typical and Atypical Development. J. Am. Acad. Child. Adolesc. Psychiatry 2020, 59, 296–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salloum-Asfar, S.; Satheesh, N.J.; Abdulla, S.A. Circulating miRNAs, Small but Promising Biomarkers for Autism Spectrum Disorder. Front. Mol. Neurosci. 2019, 12, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, W.; Zhou, S.; Zhou, J.; Wang, X. Identification of a robust non-coding RNA signature in diagnosing autism spectrum disorder by cross-validation of microarray data from peripheral blood samples. Medicine 2020, 99, e19484. [Google Scholar] [CrossRef]
- Persico, A.M.; Bourgeron, T. Searching for ways out of the autism maze: Genetic, epigenetic and environmental clues. Trends Neurosci. 2006, 29, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Loke, Y.J.; Hannan, A.J.; Craig, J. The Role of Epigenetic Change in Autism Spectrum Disorders. Front. Neurol. 2015, 6, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.C.Y.; Smith, R.G.; Hannon, E.J.; Ramaswami, G.; Parikshak, N.N.; Assary, E.; Troakes, C.; Poschmann, J.; Schalkwyk, L.C.; Sun, W.; et al. Genome-wide DNA methylation profiling identifies convergent molecular signatures associated with idiopathic and syndromic autism in post-mortem human brain tissue. Hum. Mol. Genet. 2019, 28, 2201–2211. [Google Scholar] [CrossRef] [Green Version]
- Aref-Eshghi, E.; Rodenhiser, D.I.; Schenkel, L.C.; Lin, H.; Skinner, C.; Ainsworth, P.; Paré, G.; Hood, R.L.; Bulman, D.E.; Kernohan, K.D.; et al. Genomic DNA Methylation Signatures Enable Concurrent Diagnosis and Clinical Genetic Variant Classification in Neurodevelopmental Syndromes. Am. J. Hum. Genet. 2018, 102, 156–174. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.; Rauch, T.A.; Pfeifer, G.P.; Hu, V.W. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J. 2010, 24, 3036–3051. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.C.Y.; Meaburn, E.L.; Ronald, A.R.; Price, T.S.; Jeffries, A.R.; Schalkwyk, L.C.; Plomin, R.; Mill, J. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol. Psychiatry 2014, 19, 495–503. [Google Scholar] [CrossRef]
- Andrews, S.V.; Sheppard, B.; Windham, G.; Schieve, L.A.; Schendel, D.; Croen, L.A.; Chopra, P.; Alisch, R.S.; Newschaffer, C.J.; Warren, S.T.; et al. Case-control meta-analysis of blood DNA methylation and autism spectrum disorder. Mol. Autism 2018, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Lyons, T.J.; Basu, A. Biomarkers in diabetes: Hemoglobin A1c, vascular and tissue markers. Transl. Res. 2012, 159, 303–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amunugama, R.; Jones, R.; Ford, M.; Allen, D. Bottom-Up Mass Spectrometry–Based Proteomics as an Investigative Analytical Tool for Discovery and Quantification of Proteins in Biological Samples. Adv. Wound Care 2013, 2, 549–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catherman, A.D.; Skinner, O.S.; Kelleher, N.L. Top Down proteomics: Facts and perspectives. Biochem. Biophys. Res. Commun. 2014, 445, 683–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Campo, M.; Jongbloed, W.; Twaalfhoven, H.A.M.; Veerhuis, R.; Blankenstein, M.; Teunissen, C.E. Facilitating the Validation of Novel Protein Biomarkers for Dementia: An Optimal Workflow for the Development of Sandwich Immunoassays. Front. Neurol. 2015, 6, 202. [Google Scholar] [CrossRef] [Green Version]
- Junaid, M.A.; Kowal, D.; Barua, M.; Pullarkat, P.S.; Brooks, S.S.; Pullarkat, R.K. Proteomic studies identified a single nucleotide polymorphism in glyoxalase I as autism susceptibility factor. Am. J. Med. Genet. 2004, 131, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Broek, J.A.; Guest, P.C.; Rahmoune, H.; Bahn, S. Proteomic analysis of post mortem brain tissue from autism patients: Evidence for opposite changes in prefrontal cortex and cerebellum in synaptic connectivity-related proteins. Mol. Autism 2014, 5, 41. [Google Scholar] [CrossRef] [Green Version]
- Oztan, O.; Garner, J.P.; Constantino, J.N.; Parker, K.J. Neonatal CSF vasopressin concentration predicts later medical record diagnoses of autism spectrum disorder. Proc. Natl. Acad. Sci. USA 2020, 117, 10609–10613. [Google Scholar] [CrossRef]
- Caldwell, H.K. Oxytocin and Vasopressin: Powerful Regulators of Social Behavior. Neuroscientist 2017, 23, 517–528. [Google Scholar] [CrossRef]
- Oztan, O.; Garner, J.P.; Partap, S.; Sherr, E.H.; Hardan, A.Y.; Farmer, C.; Thurm, A.; Swedo, S.E.; Parker, K.J. Cerebrospinal fluid vasopressin and symptom severity in children with autism. Ann. Neurol. 2018, 84, 611–615. [Google Scholar] [CrossRef]
- Carson, D.S.; Howerton, C.L.; Garner, J.P.; Hyde, S.A.; Clark, C.L.; Hardan, A.Y.; Penn, A.A.; Parker, K.J. Plasma vasopressin concentrations positively predict cerebrospinal fluid vasopressin concentrations in human neonates. Peptides 2014, 61, 12–16. [Google Scholar] [CrossRef]
- Wetie, A.G.N.; Wormwood, K.L.; Russell, S.; Ryan, J.P.; Darie, C.C.; Woods, A.G. A Pilot Proteomic Analysis of Salivary Biomarkers in Autism Spectrum Disorder. Autism Res. 2015, 8, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Chen, Y.; Pan, J.; Yang, A.; Niu, L.; Min, J.; Meng, X.; Liao, L.; Zhang, K.; Shen, L. Redox proteomic identification of carbonylated proteins in autism plasma: Insight into oxidative stress and its related biomarkers in autism. Clin. Proteom. 2017, 14, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Chen, Y.; Xiong, X.; Zhou, X.; Han, L.; Ni, L.; Wang, W.; Wang, X.; Zhao, L.; Shao, D.; et al. Peptidome Analysis Reveals Novel Serum Biomarkers for Children with Autism Spectrum Disorder in China. Proteom. Clin. Appl. 2018, 12, e1700164. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Du, H.-Y.; Shi, Z.-Y.; He, L.; He, Y.-Y.; Wang, D. Serum proteomic profiling for autism using magnetic bead-assisted matrix-assisted laser desorption ionization time-of-flight mass spectrometry: A pilot study. World J. Pediatr. 2018, 14, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Wetie, A.G.N.; Wormwood, K.; Thome, J.; Dudley, E.; Taurines, R.; Gerlach, M.; Woods, A.G.; Darie, C.C. A pilot proteomic study of protein markers in autism spectrum disorder. Electrophoresis 2014, 35, 2046–2054. [Google Scholar] [CrossRef] [PubMed]
- Steeb, H.; Ramsey, J.M.; Guest, P.C.; Stocki, P.; Cooper, J.D.; Rahmoune, H.; Ingudomnukul, E.; Auyeung, B.; Ruta, L.; Baron-Cohen, S.; et al. Serum proteomic analysis identifies sex-specific differences in lipid metabolism and inflammation profiles in adults diagnosed with Asperger syndrome. Mol. Autism 2014, 5, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbett, B.A.; Kantor, A.B.; Schulman, H.; Walker, W.L.; Lit, L.; Ashwood, P.; Rocke, D.M.; Sharp, F.R. A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Mol. Psychiatry 2006, 12, 292–306. [Google Scholar] [CrossRef]
- Wetie, A.G.N.; Wormwood, K.L.; Charette, L.; Ryan, J.P.; Woods, A.G.; Darie, C.C. Comparative two-dimensional polyacrylamide gel electrophoresis of the salivary proteome of children with autism spectrum disorder. J. Cell. Mol. Med. 2015, 19, 2664–2678. [Google Scholar] [CrossRef]
- Castagnola, M.; Messana, I.; Inzitari, R.; Fanali, C.; Cabras, T.; Morelli, A.; Pecoraro, A.M.; Neri, G.; Torrioli, M.G.; Gurrieri, F. Hypo-Phosphorylation of Salivary Peptidome as a Clue to the Molecular Pathogenesis of Autism Spectrum Disorders. J. Proteome Res. 2008, 7, 5327–5332. [Google Scholar] [CrossRef]
- Suganya, V.; Geetha, A.; Sujatha, S. Urine proteome analysis to evaluate protein biomarkers in children with autism. Clin. Chim. Acta 2015, 450, 210–219. [Google Scholar] [CrossRef]
- Gangadharan, A.; Nyirenda, T.; Patel, K.; Jaimes-Delgadillo, N.; Coletta, D.; Tanaka, T.; Walland, A.C.; Jameel, Z.; Vedantam, S.; Tang, S.; et al. Prolactin Induced Protein (PIP) is a potential biomarker for early stage and malignant breast cancer. Breast 2018, 39, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, A.; Jablonska, K.; Podhorska-Okolow, M.; Ugorski, M.; Dziegiel, P. Prolactin-induced protein (PIP)-characterization and role in breast cancer progression. Am. J. Cancer Res. 2018, 8, 2150–2164. [Google Scholar] [PubMed]
- Schwarz, E.; Guest, P.C.; Rahmoune, H.; Wang, L.; Levin, Y.; Ingudomnukul, E.; Ruta, L.; Kent, L.; Spain, M.; Baron-Cohen, S.; et al. Sex-specific serum biomarker patterns in adults with Asperger’s syndrome. Mol. Psychiatry 2011, 16, 1213–1220. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chauhan, A.; Sheikh, A.M.; Patil, S.; Chauhan, V.; Li, X.M.; Ji, L.; Brown, T.; Malik, M. Elevated immune response in the brain of autistic patients. J. Neuroimmunol. 2009, 207, 111–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chez, M.; Dowling, T.; Patel, P.B.; Khanna, P.; Kominsky, M. Elevation of Tumor Necrosis Factor-Alpha in Cerebrospinal Fluid of Autistic Children. Pediatric Neurol. 2007, 36, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Ashwood, P.; Krakowiak, P.; Hertz-Picciotto, I.; Hansen, R.; Pessah, I.N.; Van De Water, J. Altered T cell responses in children with autism. Brain Behav. Immun. 2011, 25, 840–849. [Google Scholar] [CrossRef] [Green Version]
- Chu, W.M. Tumor necrosis factor. Cancer Lett. 2013, 328, 222–225. [Google Scholar] [CrossRef] [Green Version]
- Naik, U.S.; Gangadharan, C.; Abbagani, K.; Nagalla, B.; Dasari, N.; Manna, S.K. A Study of Nuclear Transcription Factor-Kappa B in Childhood Autism. PLoS ONE 2011, 6, e19488. [Google Scholar] [CrossRef]
- Croonenberghs, J.; Bosmans, E.; Deboutte, D.; Kenis, G.; Maes, M. Activation of the Inflammatory Response System in Autism. Neuropsychobiology 2002, 45, 1–6. [Google Scholar] [CrossRef]
- Goines, P.E.; Croen, L.A.; Braunschweig, D.; Yoshida, C.K.; Grether, J.; Hansen, R.; Kharrazi, M.; Ashwood, P.; Van De Water, J.A. Increased midgestational IFN-γ, IL-4 and IL-5 in women bearing a child with autism: A case-control study. Mol. Autism 2011, 2, 13. [Google Scholar] [CrossRef] [Green Version]
- Tau, G.; Rothman, P. Biologic functions of the IFN-gamma receptors. Allergy 1999, 54, 1233–1251. [Google Scholar] [CrossRef] [PubMed]
- Jyonouchi, H.; Sun, S.; Le, H. Proinflammatory and regulatory cytokine production associated with innate and adaptive immune responses in children with autism spectrum disorders and developmental regression. J. Neuroimmunol. 2001, 120, 170–179. [Google Scholar] [CrossRef]
- Choi, G.B.; Yim, Y.S.; Wong, H.; Kim, S.; Kim, H.; Hoeffer, C.A.; Littman, D.R.; Huh, J.R. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 2016, 351, 933–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burré, J. The Synaptic Function of α-Synuclein. J. Parkinsons Dis. 2015, 5, 699–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellani, S.; Sousa, V.L.; Ronzitti, G.; Valtorta, F.; Meldolesi, J.; Chieregatti, E. The regulation of synaptic function by α-synuclein. Commun. Integr. Biol. 2010, 3, 106–109. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Vivacqua, G.; Yu, S. The role of α-synuclein in neurotransmission and synaptic plasticity. J. Chem. Neuroanat. 2011, 42, 242–248. [Google Scholar] [CrossRef]
- Cetin, I.; Tarakçıoğlu, M.C.; Özer, Ö.F.; Kaçar, S.; Çimen, B.; Kadak, M.T. Low Serum Level α-Synuclein and Tau Protein in Autism Spectrum Disorder Compared to Controls. Neuropediatrics 2015, 46, 410–415. [Google Scholar] [CrossRef]
- Sriwimol, W.; Limprasert, P. Significant Changes in Plasma Alpha-Synuclein and Beta-Synuclein Levels in Male Children with Autism Spectrum Disorder. BioMed Res. Int. 2018, 2018, 4503871. [Google Scholar] [CrossRef] [Green Version]
- Abou-Donia, M.B.; Suliman, H.B.; Siniscalco, D.; Antonucci, N.; Elkafrawy, P. De novo Blood Biomarkers in Autism: Autoantibodies against Neuronal and Glial Proteins. Behav. Sci. 2019, 9, 47. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, K.; Liu, F.; Gong, C.-X.; Grundke-Iqbal, I. Tau in Alzheimer Disease and Related Tauopathies. Curr. Alzheimer Res. 2010, 7, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.M.; King, J.J.; West, P.R.; Ludwig, M.A.; Donley, E.L.; Burrier, R.E.; Amaral, D.G. Amino Acid Dysregulation Metabotypes: Potential Biomarkers for Diagnosis and Individualized Treatment for Subtypes of Autism Spectrum Disorder. Biol. Psychiatry 2019, 85, 345–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, P.R.; Amaral, D.G.; Bais, P.; Smith, A.M.; Egnash, L.A.; Ross, M.E.; Palmer, J.A.; Fontaine, B.R.; Conard, K.R.; Corbett, B.A.; et al. Metabolomics as a Tool for Discovery of Biomarkers of Autism Spectrum Disorder in the Blood Plasma of Children. PLoS ONE 2014, 9, e112445. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, M.; Beck, S.E.; Haddad, S.A.; Seller, M.J. Permeability of the blood-cerebrospinal fluid barrier to plasma proteins during foetal and perinatal life. Nature 1976, 259, 140–141. [Google Scholar] [CrossRef] [PubMed]
- Atladóttir, H.Ó.; Pedersen, M.G.; Thorsen, P.; Mortensen, P.B.; Deleuran, B.; Eaton, W.W.; Parner, E.T.; Sutton, R.M.; Niles, D.; Nysaether, J.; et al. Association of Family History of Autoimmune Diseases and Autism Spectrum Disorders. Pediatrics 2009, 124, 687–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Ding, Y.; Wu, F.; Li, R.; Xie, G.; Hou, J.; Mao, P. Family history of autoimmune diseases is associated with an increased risk of autism in children: A systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2015, 55, 322–332. [Google Scholar] [CrossRef]
- Singer, H.S.; Morris, C.; Gause, C.; Pollard, M.; Zimmerman, A.W.; Pletnikov, M. Prenatal exposure to antibodies from mothers of children with autism produces neurobehavioral alterations: A pregnant dam mouse model. J. Neuroimmunol. 2009, 211, 39–48. [Google Scholar] [CrossRef]
- Brimberg, L.; Mader, S.; Jeganathan, V.; Berlin, R.; Coleman, T.R.; Gregersen, P.K.; Huerta, P.T.; Volpe, B.T.; Diamond, B. Caspr2-reactive antibody cloned from a mother of an ASD child mediates an ASD-like phenotype in mice. Mol. Psychiatry 2016, 21, 1663–1671. [Google Scholar] [CrossRef] [Green Version]
- Braunschweig, D.; Duncanson, P.; Boyce, R.; Hansen, R.; Ashwood, P.; Pessah, I.N.; Hertz-Picciotto, I.; Van De Water, J.A. Behavioral Correlates of Maternal Antibody Status Among Children with Autism. J. Autism Dev. Disord. 2011, 42, 1435–1445. [Google Scholar] [CrossRef] [Green Version]
- Croen, L.A.; Braunschweig, D.; Haapanen, L.; Yoshida, C.K.; Fireman, B.; Grether, J.K.; Kharrazi, M.; Hansen, R.L.; Ashwood, P.; Van De Water, J. Maternal Mid-Pregnancy Autoantibodies to Fetal Brain Protein: The Early Markers for Autism Study. Biol. Psychiatry 2008, 64, 583–588. [Google Scholar] [CrossRef] [Green Version]
- Braunschweig, D.; Ashwood, P.; Krakowiak, P.; Hertz-Picciotto, I.; Hansen, R.; Croen, L.A.; Pessah, I.N.; Van De Water, J.A. Autism: Maternally derived antibodies specific for fetal brain proteins. NeuroToxicology 2007, 29, 226–231. [Google Scholar] [CrossRef] [Green Version]
- Braunschweig, D.; Krakowiak, P.; Duncanson, P.; Boyce, R.; Hansen, R.L.; Ashwood, P.; Hertz-Picciotto, I.; Pessah, I.N.; Van De Water, J.A. Autism-specific maternal autoantibodies recognize critical proteins in developing brain. Transl. Psychiatry 2013, 3, e277. [Google Scholar] [CrossRef] [PubMed]
- Heuer, L.; Braunschweig, D.; Ashwood, P.; Van De Water, J.A.; Campbell, D.B. Association of a MET genetic variant with autism-associated maternal autoantibodies to fetal brain proteins and cytokine expression. Transl. Psychiatry 2011, 1, e48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singer, H.S.; Morris, C.M.; Gause, C.D.; Gillin, P.K.; Crawford, S.; Zimmerman, A.W. Antibodies against fetal brain in sera of mothers with autistic children. J. Neuroimmunol. 2008, 194, 165–172. [Google Scholar] [CrossRef]
- Shen, L.; Liu, X.; Zhang, H.; Lin, J.; Feng, C.; Iqbal, J. Biomarkers in autism spectrum disorders: Current progress. Clin. Chim. Acta 2020, 502, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.-M.; Sepers, M.; Henstridge, C.M.; Lassalle, O.; Neuhofer, D.; Martin, H.; Ginger, M.; Frick, A.; DiPatrizio, N.V.; Mackie, K.; et al. Uncoupling of the endocannabinoid signalling complex in a mouse model of fragile X syndrome. Nat. Commun. 2012, 3, 1080. [Google Scholar] [CrossRef] [Green Version]
- Földy, C.; Malenka, R.C.; Südhof, T.C. Autism-Associated Neuroligin-3 Mutations Commonly Disrupt Tonic Endocannabinoid Signaling. Neuron 2013, 78, 498–509. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Dinh, D.; Lee, D.; Allison, A.; Anguren, A.; Moreno-Sanz, G.; Gall, C.M.; Piomelli, D. Enhancement of Anandamide-Mediated Endocannabinoid Signaling Corrects Autism-Related Social Impairment. Cannabis Cannabinoid Res. 2016, 1, 81–89. [Google Scholar] [CrossRef]
- Kerr, D.; Downey, L.; Conboy, M.; Finn, D.; Roche, M. Alterations in the endocannabinoid system in the rat valproic acid model of autism. Behav. Brain Res. 2013, 249, 124–132. [Google Scholar] [CrossRef]
- Karhson, D.S.; Krasinska, K.M.; Ahloy-Dallaire, J.; Libove, R.A.; Phillips, J.M.; Chien, A.S.; Garner, J.P.; Hardan, A.Y.; Parker, K.J. Plasma anandamide concentrations are lower in children with autism spectrum disorder. Mol. Autism 2018, 9, 18. [Google Scholar] [CrossRef]
- Aran, A.; Eylon, M.; Harel, M.; Polianski, L.; Nemirovski, A.; Tepper, S.; Schnapp, A.; Ecassuto, H.; Wattad, N.; Tam, J. Lower circulating endocannabinoid levels in children with autism spectrum disorder. Mol. Autism 2019, 10, 2. [Google Scholar] [CrossRef]
- Servadio, M.; Melancia, F.; Manduca, A.; Di Masi, A.; Schiavi, S.; Cartocci, V.; Pallottini, V.; Campolongo, P.; Ascenzi, P.; Trezza, V. Targeting anandamide metabolism rescues core and associated autistic-like symptoms in rats prenatally exposed to valproic acid. Transl. Psychiatry 2016, 6, e902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Otaish, H.; Al-Ayadhi, L.Y.; Bjørklund, G.; Chirumbolo, S.; Urbina, M.A.; El-Ansary, A. Relationship between absolute and relative ratios of glutamate, glutamine and GABA and severity of autism spectrum disorder. Metab. Brain Dis. 2018, 33, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, C.F.; Puglia, M.P.; Zucca, S. Focus On: Neurotransmitter Systems. Alcohol Res. Health J. Natl. Inst. Alcohol Abus. Alcohol. 2011, 34, 106–120. [Google Scholar]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.W.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism—Comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef] [Green Version]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids from Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [Green Version]
- Oh, D.; Cheon, K.-A. Alteration of Gut Microbiota in Autism Spectrum Disorder: An Overview. J. Korean Acad. Child. Adolesc. Psychiatry 2020, 31, 131–145. [Google Scholar] [CrossRef]
- Fulceri, F.; Morelli, M.; Santocchi, E.; Cena, H.; Del Bianco, T.; Narzisi, A.; Calderoni, S.; Muratori, F. Gastrointestinal symptoms and behavioral problems in preschoolers with Autism Spectrum Disorder. Dig. Liver Dis. 2016, 48, 248–254. [Google Scholar] [CrossRef]
- McElhanon, B.O.; McCracken, C.; Karpen, S.; Sharp, W.G. Gastrointestinal Symptoms in Autism Spectrum Disorder: A Meta-analysis. Pediatrics 2014, 133, 872–883. [Google Scholar] [CrossRef] [Green Version]
- Chaidez, V.; Hansen, R.L.; Hertz-Picciotto, I. Gastrointestinal Problems in Children with Autism, Developmental Delays or Typical Development. J. Autism Dev. Disord. 2014, 44, 1117–1127. [Google Scholar] [CrossRef] [Green Version]
- Rao, M.; Gershon, M.D. The bowel and beyond: The enteric nervous system in neurological disorders. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 517–528. [Google Scholar] [CrossRef] [Green Version]
- Bernier, R.; Golzio, C.; Xiong, B.; Stessman, H.A.; Coe, B.P.; Penn, O.; Witherspoon, K.; Gerdts, J.; Baker, C.; Vulto-van Silfhout, A.T.; et al. Disruptive CHD8 Mutations Define a Subtype of Autism Early in Development. Cell 2014, 158, 263–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharon, G.; Cruz, N.J.; Kang, D.-W.; Gandal, M.J.; Wang, B.; Kim, Y.-M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 2019, 177, 1600–1618.e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.-W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; LaBaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced Incidence of Prevotella and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, B.L.; Hornig, M.; Buie, T.; Bauman, M.L.; Paik, M.C.; Wick, I.; Bennett, A.; Jabado, O.; Hirschberg, D.L.; Lipkin, W.I. Impaired Carbohydrate Digestion and Transport and Mucosal Dysbiosis in the Intestines of Children with Autism and Gastrointestinal Disturbances. PLoS ONE 2011, 6, e24585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parracho, H.M.R.T.; Bingham, M.O.; Gibson, G.R.; McCartney, A.L. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J. Med. Microbiol. 2005, 54, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Shaw, W. Increased urinary excretion of a 3-(3-hydroxyphenyl)-3-hydroxypropionic acid (HPHPA), an abnormal phenylalanine metabolite ofClostridiaspp. in the gastrointestinal tract, in urine samples from patients with autism and schizophrenia. Nutr. Neurosci. 2010, 13, 135–143. [Google Scholar] [CrossRef]
- Altieri, L.; Neri, C.; Sacco, R.; Curatolo, P.; Benvenuto, A.; Muratori, F.; Santocchi, E.; Bravaccio, C.; Lenti, C.; Saccani, M.; et al. Urinary p-cresol is elevated in small children with severe autism spectrum disorder. Biomarkers 2011, 16, 252–260. [Google Scholar] [CrossRef]
- Gabriele, S.; Sacco, R.; Cerullo, S.; Neri, C.; Urbani, A.; Tripi, G.; Malvy, J.; Barthelemy, C.; Bonnet-Brihault, F.; Persico, A.M. Urinary p-cresol is elevated in young French children with autism spectrum disorder: A replication study. Biomarkers 2014, 19, 463–470. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [Green Version]
- Passmore, I.J.; Letertre, M.P.; Preston, M.D.; Bianconi, I.; Harrison, M.A.; Nasher, F.; Kaur, H.; Hong, H.A.; Baines, S.D.; Cutting, S.M.; et al. Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria. PLOS Pathog. 2018, 14, e1007191. [Google Scholar] [CrossRef] [Green Version]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santocchi, E.; Guiducci, L.; Prosperi, M.; Calderoni, S.; Gaggini, M.; Apicella, F.; Tancredi, R.; Billeci, L.; Mastromarino, P.; Grossi, E.; et al. Effects of Probiotic Supplementation on Gastrointestinal, Sensory and Core Symptoms in Autism Spectrum Disorders: A Randomized Controlled Trial. Front. Psychiatry 2020, 11, 550593. [Google Scholar] [CrossRef] [PubMed]
- Hassan, W.M.; Al-Ayadhi, L.Y.; Bjørklund, G.; Alabdali, A.; Chirumbolo, S.; El-Ansary, A. The Use of Multi-parametric Biomarker Profiles May Increase the Accuracy of ASD Prediction. J. Mol. Neurosci. 2018, 66, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, D.; Frye, R.E. Mitochondrial dysfunction in autism spectrum disorders: A systematic review and meta-analysis. Mol. Psychiatry 2011, 17, 290–314. [Google Scholar] [CrossRef] [Green Version]
- Singh, K.; Singh, I.N.; Diggins, E.; Connors, S.L.; Karim, M.A.; Lee, D.; Zimmerman, A.W.; Frye, R.E. Developmental regression and mitochondrial function in children with autism. Ann. Clin. Transl. Neurol. 2020, 7, 683–694. [Google Scholar] [CrossRef]
- Giulivi, C.; Zhang, Y.-F.; Omanska-Klusek, A.; Ross-Inta, C.; Wong, S.; Hertz-Picciotto, I.; Tassone, F.; Pessah, I.N. Mitochondrial Dysfunction in Autism. JAMA 2010, 304, 2389–2396. [Google Scholar] [CrossRef] [Green Version]
- Shoffner, J.; Hyams, L.; Langley, G.N.; Cossette, S.; Mylacraine, L.; Dale, J.; Ollis, L.; Kuoch, S.; Bennett, K.; Aliberti, A.; et al. Fever Plus Mitochondrial Disease Could Be Risk Factors for Autistic Regression. J. Child. Neurol. 2009, 25, 429–434. [Google Scholar] [CrossRef]
- Chaudhari, N.; Talwar, P.; Parimisetty, A.; Lefebvre d’Hellencourt, C.; Ravanan, P. A molecular web: Endoplasmic reticulum stress, inflammation, and oxidative stress. Front. Cell. Neurosci. 2014, 8, 213. [Google Scholar] [CrossRef]
- Libero, L.E.; Nordahl, C.W.; Li, D.D.; Ferrer, E.; Rogers, S.J.; Amaral, D.G. Persistence of megalencephaly in a subgroup of young boys with autism spectrum disorder. Autism Res. 2016, 9, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- Nordahl, C.W.; Braunschweig, D.; Iosif, A.-M.; Lee, A.; Rogers, S.; Ashwood, P.; Amaral, D.G.; Van De Water, J. Maternal autoantibodies are associated with abnormal brain enlargement in a subgroup of children with autism spectrum disorder. Brain Behav. Immun. 2013, 30, 61–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, M.; Lombardo, M.V.; Auyeung, B.; Chakrabarti, B.; Baron-Cohen, S. Sex/Gender Differences and Autism: Setting the Scene for Future Research. J. Am. Acad. Child. Adolesc. Psychiatry 2015, 54, 11–24. [Google Scholar] [CrossRef] [Green Version]
- Foster, N.E.; Doyle-Thomas, K.A.; Tryfon, A.; Ouimet, T.; Anagnostou, E.; Evans, A.C.; Zwaigenbaum, L.; Lerch, J.P.; Lewis, J.D.; Hyde, K.L. Structural Gray Matter Differences During Childhood Development in Autism Spectrum Disorder: A Multimetric Approach. Pediatric Neurol. 2015, 53, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Haar, S.; Berman, S.; Behrmann, M.; Dinstein, I. Anatomical Abnormalities in Autism? Cereb. Cortex 2014, 26, 1440–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ure, A.M.; Treyvaud, K.; Thompson, D.K.; Pascoe, L.; Roberts, G.; Lee, K.J.; Seal, M.L.; Northam, E.; Cheong, J.L.; Hunt, R.W.; et al. Neonatal brain abnormalities associated with autism spectrum disorder in children born very preterm. Autism Res. 2015, 9, 543–552. [Google Scholar] [CrossRef]
- Brun, C.C.; Nicolson, R.; Leporé, N.; Chou, Y.-Y.; Vidal, C.N.; DeVito, T.J.; Drost, D.J.; Williamson, P.C.; Rajakumar, N.; Toga, A.W.; et al. Mapping brain abnormalities in boys with autism. Hum. Brain Mapp. 2009, 30, 3887–3900. [Google Scholar] [CrossRef] [Green Version]
- Schumann, C.M.; Barnes, C.C.; Lord, C.; Courchesne, E. Amygdala Enlargement in Toddlers with Autism Related to Severity of Social and Communication Impairments. Biol. Psychiatry 2009, 66, 942–949. [Google Scholar] [CrossRef] [Green Version]
- Schumann, C.M.; Hamstra, J.; Goodlin-Jones, B.L.; Lotspeich, L.J.; Kwon, H.; Buonocore, M.H.; Lammers, C.R.; Reiss, A.L.; Amaral, D.G. The Amygdala Is Enlarged in Children but Not Adolescents with Autism; the Hippocampus Is Enlarged at All Ages. J. Neurosci. 2004, 24, 6392–6401. [Google Scholar] [CrossRef]
- Shen, M.D.; Nordahl, C.W.; Young, G.S.; Wootton-Gorges, S.L.; Lee, A.; Liston, S.E.; Harrington, K.R.; Ozonoff, S.; Amaral, D.G. Early brain enlargement and elevated extra-axial fluid in infants who develop autism spectrum disorder. Brain 2013, 136, 2825–2835. [Google Scholar] [CrossRef] [Green Version]
- Emerson, R.W.; Adams, C.; Nishino, T.; Hazlett, H.C.; Wolff, J.J.; Zwaigenbaum, L.; Constantino, J.N.; Shen, M.D.; Swanson, M.R.; Elison, J.T.; et al. Functional neuroimaging of high-risk 6-month-old infants predicts a diagnosis of autism at 24 months of age. Sci. Transl. Med. 2017, 9, eaag2882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, G.; Carver, L.; Meltzoff, A.N.; Panagiotides, H.; McPartland, J.; Webb, S.J. Neural Correlates of Face and Object Recognition in Young Children with Autism Spectrum Disorder, Developmental Delay, and Typical Development. Child. Dev. 2002, 73, 700–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagnozzi, A.M.; Conti, E.; Calderoni, S.; Fripp, J.; Rose, S.E. A systematic review of structural MRI biomarkers in autism spectrum disorder: A machine learning perspective. Int. J. Dev. Neurosci. 2018, 71, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.; Martin, T.; Downie, J.; Malisza, K. A Comprehensive Analysis of MRI Research Risks: In Support of Full Disclosure. Can. J. Neurol. Sci. 2007, 34, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Falck-Ytter, T.; Bölte, S.; Gredebäck, G. Eye tracking in early autism research. J. Neurodev. Disord. 2013, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Falck-Ytter, T.; Fernell, E.; Hedvall, Å.L.; Von Hofsten, C.; Gillberg, C. Gaze Performance in Children with Autism Spectrum Disorder when Observing Communicative Actions. J. Autism Dev. Disord. 2012, 42, 2236–2245. [Google Scholar] [CrossRef] [PubMed]
- Dalton, K.M.; Nacewicz, B.M.; Johnstone, T.; Schaefer, H.S.; Gernsbacher, M.A.; Goldsmith, H.H.; Alexander, A.L.; Davidson, R.J. Gaze fixation and the neural circuitry of face processing in autism. Nat. Neurosci. 2005, 8, 519–526. [Google Scholar] [CrossRef] [Green Version]
- Klin, A.; Jones, W.; Schultz, R.; Volkmar, F.; Cohen, D. Visual Fixation Patterns during Viewing of Naturalistic Social Situations as Predictors of Social Competence in Individuals with Autism. Arch. Gen. Psychiatry 2002, 59, 809–816. [Google Scholar] [CrossRef] [Green Version]
- Wan, G.; Kong, X.; Sun, B.; Yu, S.; Tu, Y.; Park, J.; Lang, C.; Koh, M.; Wei, Z.; Feng, Z.; et al. Applying Eye Tracking to Identify Autism Spectrum Disorder in Children. J. Autism Dev. Disord. 2019, 49, 209–215. [Google Scholar] [CrossRef]
- Jones, W.; Carr, K.; Klin, A. Absence of Preferential Looking to the Eyes of Approaching Adults Predicts Level of Social Disability in 2-Year-Old Toddlers with Autism Spectrum Disorder. Arch. Gen. Psychiatry 2008, 65, 946–954. [Google Scholar] [CrossRef] [Green Version]
- Loth, E.; Murphy, D.G.; Spooren, W. Defining Precision Medicine Approaches to Autism Spectrum Disorders: Concepts and Challenges. Front. Psychiatry 2016, 7, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beversdorf, D.Q. Phenotyping, Etiological Factors, and Biomarkers: Toward Precision Medicine in Autism Spectrum Disorders. J. Dev. Behav. Pediatrics 2016, 37, 659–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, E.H.; Zabner, J. Precision Genomic Medicine in Cystic Fibrosis. Clin. Transl. Sci. 2015, 8, 606–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLean, E.; Cogswell, M.; Egli, I.; Wojdyla, D.; De Benoist, B. Worldwide prevalence of anaemia, WHO Vitamin and Mineral Nutrition Information System, 1993–2005. Public Health Nutr. 2008, 12, 444–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Northrop-Clewes, C.A.; Thurnham, D.I. Biomarkers for the differentiation of anemia and their clinical usefulness. J. Blood Med. 2013, 4, 11–22. [Google Scholar]
- Mitsudomi, T.; Suda, K.; Yatabe, Y. Surgery for NSCLC in the era of personalized medicine. Nat. Rev. Clin. Oncol. 2013, 10, 235–244. [Google Scholar] [CrossRef]
- Ginsburg, G.S.; Phillips, K.A. Precision Medicine: From Science to Value. Health Aff. 2018, 37, 694–701. [Google Scholar] [CrossRef]
- Dainis, A.M.; Ashley, E. Cardiovascular Precision Medicine in the Genomics Era. JACC Basic Transl. Sci. 2018, 3, 313–326. [Google Scholar] [CrossRef]
- Luo, Y.; Eran, A.; Palmer, N.; Avillach, P.; Levy-Moonshine, A.; Szolovits, P.; Kohane, I.S. A multidimensional precision medicine approach identifies an autism subtype characterized by dyslipidemia. Nat. Med. 2020, 26, 1375–1379. [Google Scholar] [CrossRef]


| Gene Name | Function | Reference |
|---|---|---|
| Astrotactin- 2 (ASTN2) | Neuronal adhesion molecule has a role in glial migration. | [19] |
| Contactin 4 (CNTN4) | Neuronal maintenance and plasticity. | [20] |
| F-Box Protein- 40 (FBXO40) | Ubiquitin-protein transferase activity. | [19] |
| FMRP translational regulator-1 (FMR1) | mRNA trafficking from the nucleus to the cytoplasm. Synaptic plasticity. | [21] |
| Potassium voltage-gated channel subfamily Q member 2 (KCNQ2) | Transports potassium ions inside and outside the cells. | [22] |
| lysine methyltransferase 2E (KMT2E) | Regulates gene transcription. | [22] |
| Mono-ADP-Ribosylhydrolase (MACROD2) | Remove ADP-ribose from mono-ADP-ribosylated proteins. | [22] |
| Methyl CpG binding protein- 2 (MeCP2) | Chromosomal protein that binds to methylated DNA, it binds to single methy-CpG pairs. | [21] |
| Neuronal growth regulator- 1 (NEGR1) | Regulates synapses formation in the hippocampus. | [22] |
| Neuroligin-1 (NLGN1) | Synaptic functions and transmission. | [19] |
| Neurexin- 1 (NRXN1) | Binds neuroligins and formation of synaptic contacts. | [23] |
| Parkin (PARK2) | Part of protease complex multiprotein that guides to proteasomal degradation. | [19] |
| Polypyrimidine tract binding protein-2 (PTBP2) | Control assembly of splicing- regulatory proteins and important for alternative splicing in early development. | [22] |
| Ring finger and WD domain 2 (RFWD2) Also, known as COP1 | Mediates ubiquitination and substrate protein degradation. | [19] |
| SH3 and multiple ankyrin repeat domains protein-3 (SHANK3) | Scaffold protein of the postsynaptic density. | [24] |
| Tuberous sclerosis complex (TSC1 and TSC2) | Tumor suppressor gene that activate GTPase activating protein tuberin. | [25] |
| ubiquitin protein ligase E3A (UBE3A) | E3 ubiquitin-protein ligase. | [19] |
| Sample Type | Detection Method | Proteins Identified | Function | Reference |
|---|---|---|---|---|
| Serum | Tricine-PAGE LC-MS/MS | ApoA1, ApoA4, PON1. | Cholesterol metabolism Oxidative damage | [55] |
| Serum | multiplex immunoassay LC-MS | Immune assays Females: ADIPO, APOA1, IgA Males: IL-12p70, IL-16, TF, TNF-alpha, BMP6, CTGF, ICAM1. Both: CHGA, EPO, IL-3, TENA, PAP, SHBG. LC-MS Females: APOC2, APOE, ARMC3, CLC4K, FETUB, GLCE, MRRP1, PTPA, RN149, TLE1, TRIPB, ZC3HE. Males: RGPD4. | Cholesterol metabolism and transport Inflammation Androgens | [56] |
| Serum | MALDI-TOF MS | SERPINA5, PF4, FABP1, APOC1, AFP, CPB2, TAAR6, FGA. | Platelets and coagulation functions Cholesterol metabolism | [53] |
| Serum | MALDI-TOF MS | Eight peaks (Unidentified) 6.42 kDa, 7.75 kDa, 9.27 kDa, 3.88 kDa, 6.62 kDa, 4.08 kDa, 4.64 kDa, 4.20 kDa. | - | [54] |
| Serum | LC-ESI-MS on TOF | FHR1, FN1, C1q, B-100. | Cholesterol metabolism Complement system | [57] |
| Saliva | Nano LC-MS | PIP, LTF, IGKC, IGHG1, IGLC2, NE, pIgR, DMBT1. | Immune system | [51] |
| Saliva | 2D-PAGE Nano LC-MS/MS | AMY1A, CBP, P532, TF, ZAG, ZG16, CST5, PLG, FRAT1, KIF14, ITGA 6, GRTP1, hPSP, PIP, MUC16. | Lipid and cholesterol metabolism Immune system Oxidative damage | [58] |
| Saliva | LC-MS/MS | Hypo-phosphorylation of STATH, HTN1, aPRP. | - | [59] |
| Urine | 2D-PAGE MALDI-TOF MS | KNG-1, MASP2, IGHG1. | Immune system Coagulation | [60] |
| Observed Autoantibodies Reactivities | Molecular Weight | Samples Used | Reference |
|---|---|---|---|
| Human fetal brain proteins Human adult brain proteins | 73 kDa and 37 kD - | Mother’s serum of: ASD vs. non-ASD (DD) * vs. TD * | [90] |
| Human fetal brain proteins | 39 kDa, 39 kDa and 73 kDa | Mother’s plasma of: AU * vs. ASD vs. DD * vs. TD * | [88] |
| Human fetal brain proteins Human adult brain proteins Rodent embryo brain proteins Rodent adult brain proteins | 36 kDa, 39 kDa and 61 kDa caudate at 155 kDa and BA9 at 63 kDa 36 kDa and 73 kDa 27 kDa | Mother’s serum of: ASD vs. controls | [93] |
| LDH 1, LDH2, STIP, CRMP1, CRMP2, and YBX1 | 37 kDa, 39 kDa, 48 kDa, 62 kDa and 68 kDa | Mother’s plasma of: ASD vs. controls | [91] |
| Metabolite | Sample Type | Method of Detection | Effect/Function | Reference |
|---|---|---|---|---|
| Anandamide (decrease) | Serum/Plasma | LC-MS/MS | Endocannabinoid signaling | [99,100] |
| HPHPA (increase) | Urine | GC-MS | A by-product of clostridium species and a probable tyrosine analog of m-tyrosine (3-hydroxyphenylalanine), that may depletes brain catecholamines. | [116] |
| p-hydroxyphenyllactat (decrease) | Plasma | LC-HRMS | A by-product of bifidobacteria and lactobacillus, act as an anti-oxidant | [82] |
| p-cresol (increase) | Urine Feces | HPLC-UV GC-MS/SPME | Competes with neurotransmitters on the sulfonation process and disturbs gut microbiome | [117,118,119] |
| GABA (increase) | Plasma | ELISA | Neurotransmitter | [102,125] |
| Glutamic acid (increase) | Plasma | LC-HRMS | Amino acid | [82] |
| SCFA (decrease) | Feces | FID-GC | Regulate tryptophan 5-hydroxylase 1 which is important for serotonin, dopamine, adrenaline and nor adrenaline production. | [104] |
| Lactate (increase) | Serum | ELISA and colorimetric assays | Energy metabolism | [125] |
| Pyruvate (increase) | Serum | ELISA and colorimetric assays | Energy metabolism | [125] |
| 5-Aminovaleric acid (increase) | Plasma | LC-HRMS | Lysine degradation product and week inhibitor of coagulation | [82] |
| DHEA-sulfate (increase) | Plasma | LC-HRMS | Sex-hormone | [82] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mesleh, A.G.; Abdulla, S.A.; El-Agnaf, O. Paving the Way toward Personalized Medicine: Current Advances and Challenges in Multi-OMICS Approach in Autism Spectrum Disorder for Biomarkers Discovery and Patient Stratification. J. Pers. Med. 2021, 11, 41. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm11010041
Mesleh AG, Abdulla SA, El-Agnaf O. Paving the Way toward Personalized Medicine: Current Advances and Challenges in Multi-OMICS Approach in Autism Spectrum Disorder for Biomarkers Discovery and Patient Stratification. Journal of Personalized Medicine. 2021; 11(1):41. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm11010041
Chicago/Turabian StyleMesleh, Areej G., Sara A. Abdulla, and Omar El-Agnaf. 2021. "Paving the Way toward Personalized Medicine: Current Advances and Challenges in Multi-OMICS Approach in Autism Spectrum Disorder for Biomarkers Discovery and Patient Stratification" Journal of Personalized Medicine 11, no. 1: 41. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm11010041