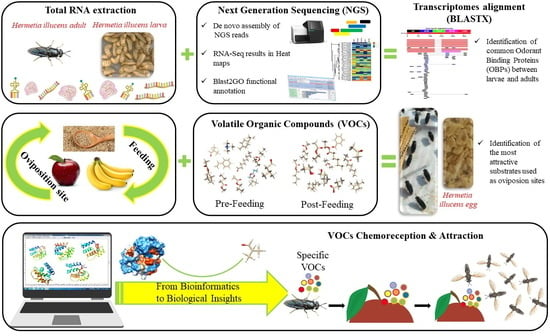
Hermetia illucens (L.) (Diptera: Stratiomyidae) Odorant Binding Proteins and Their Interactions with Selected Volatile Organic Compounds: An In Silico Approach
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. RNA Extraction from BSF Larvae and Adults
2.3. RNA Sequencing and Transcriptome De Novo Assembly
2.4. Identification of Chemosensory Genes
2.5. Differential Expression of OBP Genes in Adult BSF and Identification of Common OBPs in Larval and Adult Transcriptome
2.6. Volatile Organic Compound (VOC) Sampling
2.7. GC/MS Analysis
2.8. Ab Initio Modelling of OBPs and Virtual Ligand Screening
3. Results
3.1. Candidate Chemosensory Genes in BSF Adult and Larval Transcriptomes
3.2. Differential Expression of OBP in BSF Adults and Larvae
3.3. Identification of Common OBPs in Larval and Adult Transcriptomes
3.4. Identification of Volatile Organic Compounds
3.5. Molecular Modelling and Virtual Docking of OBPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.S.; Shelomi, M. Review of black soldier fly (Hermetia illucens) as animal feed and human food. Foods 2017, 6, 91. [Google Scholar] [CrossRef] [Green Version]
- Diener, S.; Zurbrügg, C.; Roa-Gutiérrez, F.; Hong Dang, N.; Morel, A.; Koottatep, T.; Tockner, K. Black soldier fly larvae for organic waste treatment–prospects and constraints. In Proceedings of the WasteSafe 2011—2nd International Conference on Solid Waste Management in the Developing Countries, Khulna, Bangladesh, 13–15 February 2011; Alamgir, M., Bari, Q.H., Rafizul, I.M., Islam, S.M.T., Sarkar, G., Howladerm, M.K., Eds.; pp. 52–59. [Google Scholar]
- Nguyen, T.T.X.; Tomberlin, J.K.; Vanlaerhoven, S. Ability of Black Soldier Fly (Diptera: Stratiomyidae) Larvae to Recycle Food Waste. Environ. Entomol. 2015, 44, 406–410. [Google Scholar] [CrossRef]
- Jucker, C.; Erba, D.; Leonardi, M.G.; Lupi, D.; Savoldelli, S. Assessment of Vegetable and Fruit Substrates as Potential Rearing Media for Hermetia illucens (Diptera: Stratiomyidae) Larvae. Environ. Entomol. 2017, 46, 1415–1423. [Google Scholar] [CrossRef]
- Salomone, R.; Saija, G.; Mondello, G.; Giannetto, A.; Fasulo, S.; Savastano, D. Environmental impact of food waste bioconversion by insects: Application of life cycle assessment to process using Hermetia illucens. J. Clean. Prod. 2017, 140, 890–905. [Google Scholar] [CrossRef]
- Sheppard, C. Housefly and lesser fly control utilizing the black soldier fly in manure management systems for caged laying hens. Environ. Entomol. 1983, 12, 1439–1442. [Google Scholar] [CrossRef]
- Popa, R.; Green, T.R. Using black soldier fly larvae for processing organic leachates. J. Econ. Entomol. 2012, 105, 374–378. [Google Scholar] [CrossRef]
- Webster, C.D.; Rawles, S.; Koch, J.; Thompson, K.; Kobayashi, Y.; Gannam, A.; Twibell, R.; Hyde, N. Bio-Ag reutilization of distiller’s dried grains with solubles (DDGS) as a substrate for black soldier fly larvae, Hermetia illucens, along with poultry by-product meal and soybean meal, as total replacement of fish meal in diets for Nile tilapia, Oreochromis niloticus. Aquacult. Nutr. 2016, 22, 976–988. [Google Scholar]
- Scala, A.; Cammack, J.A.; Salvia, R.; Scieuzo, C.; Franco, A.; Bufo, S.A.; Tomberlin, J.K.; Falabella, P. Rearing substrate impacts growth and macronutrient composition of Hermetia illucens (L.) (Diptera: Stratiomyidae) larvae produced at an industrial scale. Sci. Rep. 2020, 10, 19448. [Google Scholar] [CrossRef]
- Spinelli, R.; Neri, P.; Pini, M.; Barbi, S.; Monia, M.; Ferrari, A. Using Black Soldier Flies (Hermetia Illucens) To Bioconvert Waste from The Livestock Production Chain: A Life Cycle Assessment Case Study. Waste Manag. Environ. IX 2018, 231, 47–58. [Google Scholar]
- Kim, W.; Bae, S.; Lee, S.; Choi, Y.; Han, S.; Koh, Y.H. Biochemical characterization of digestive enzymes in the black soldier fly, Hermetia illucens (diptera: Stratiomyidae). J. Asia Pac. Entomol. 2011, 14, 11–14. [Google Scholar] [CrossRef]
- Pezzi, M.; Scapoli, C.; Bharti, M.; Faucheux, M.J.; Chicca, M.; Leis, M.; Marchetti, M.G.; Mamolini, E.; Salvia, R.; Falabella, P.; et al. Fine Structure of Maxillary Palps in Adults of Hermetia illucens (Diptera: Stratiomyidae). J. Med. Entomol. 2021, 58, 658–665. [Google Scholar] [CrossRef]
- Lalander, C.; Diener, S.; Magri, M.E.; Zurbrügg, C.; Lindström, A.; Vinnerås, B. Faecal sludge management with the larvae of the black soldier fly (Hermetia illucens)—From a hygiene aspect. Sci. Total. Environ. 2013, 458, 312–318. [Google Scholar] [CrossRef]
- Zhou, F.; Tomberlin, J.K.; Zheng, L.; Yu, Z.; Zhang, J. Developmental and waste reduction plasticity of three black soldier fly strains (Diptera: Stratiomyidae) raised on different livestock manures. J. Med. Entomol. 2013, 50, 1224–1230. [Google Scholar] [CrossRef] [PubMed]
- Erickson, M.C.; Islam, M.; Sheppard, C.; Liao, J.; Doyle, M.P. Reduction of Escherichia coli O157:H7 and Salmonella enterica serovar Enteritidis in chicken manure by larvae of the black soldier fly. J. Food Prot. 2004, 67, 685–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Tomberlin, J.K.; Brady, J.A.; Sanford, M.R.; Yu, Z. Black soldier fly (Diptera: Stratiomyidae) larvae reduce Escherichia coli in dairy manure. Environ. Entomol. 2008, 37, 1525–1530. [Google Scholar] [CrossRef]
- Moretta, A.; Salvia, R.; Scieuzo, C.; Di Somma, A.; Vogel, H.; Pucci, P.; Sgambato, A.; Wolff, M.; Falabella, P. A bioinformatic study of antimicrobial peptides identified in the Black Soldier Fly (BSF) Hermetia illucens (Diptera: Stratiomyidae). Sci. Rep. 2020, 10, 16875. [Google Scholar] [CrossRef]
- Manniello, M.D.; Moretta, A.; Salvia, R.; Scieuzo, C.; Lucchetti, D.; Vogel, H.; Sgambato, A.; Falabella, P. Insect antimicrobial peptides: Potential weapons to counteract the antibiotic resistance. Cell. Mol. Life Sci. 2021, 78, 4259–4282. [Google Scholar] [CrossRef]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell. Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef]
- Lima, E.; Ferreira, C.P.; Bernardes, A.M.; Godoy, W.A.C. Neighborhood interactions and larval dispersal behavior in blowflies. J. Insect Behav. 2009, 22, 245–255. [Google Scholar] [CrossRef]
- Hoc, B.; Noël, G.; Carpentier, J.; Francis, F.; Caparros Megido, R. Optimization of black soldier fly (Hermetia illucens) artificial reproduction. PLoS ONE 2019, 14, e0216160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomberlin, J.K.; Adler, P.A.; Myers, H.M. Development of the Black Soldier Fly (Diptera: Stratiomyidae) in Relation to Temperature. Environm. Entomol. 2009, 38, 930–934. [Google Scholar] [CrossRef]
- Holmes, L.A.; Vanlaerhoven, S.L.; Tomberlin, J.K. Relative Humidity Effects on the Life History of Hermetia illucens (Diptera: Stratiomyidae). Environ. Entomol. 2012, 41, 971–978. [Google Scholar] [CrossRef] [Green Version]
- Heussler, C.D.; Walter, A.; Oberkofler, H.; Insam, H.; Arthofer, W.; Schlick-Steiner, B.C.; Steiner, F.M. Influence of three artificial light sources on oviposition and half-life of the Black Soldier Fly, Hermetia illucens (Diptera: Stratiomyidae): Improving small-scale indoor rearing. PLoS ONE 2018, 13, e0197896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguiar, J.; Gonçalves, J.L.; Alves, V.L.; Câmara, J.S. Relationship between Volatile Composition and Bioactive Potential of Vegetables and Fruits of Regular Consumption—An Integrative Approach. Molecules 2021, 26, 3653. [Google Scholar] [CrossRef]
- Jørgensen, L.V.; Huss, H.H.; Dalgaard, P. Significance of volatile compounds produced by spoilage bacteria in vacuum-packed cold-smoked salmon (Salmo salar) analyzed by GC-MS and multivariate regression. J. Agric. Food Chem. 2001, 49, 2376–2381. [Google Scholar] [CrossRef] [PubMed]
- Parlapani, F.F.; Haroutounian, S.A.; Nychas, G.J.E.; Boziaris, I.S. Microbiological spoilage and volatiles production of gutted European sea bass stored under air and commercial modified atmosphere package at 2 °C. Food Microbiol. 2015, 50, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Xie, J. Growth kinetics and spoilage potential of co-culturing Acinetobacter johnsonii and Pseudomonas fluorescens from bigeye tuna (Thunnus obesus) during refrigerated storage. Curr. Microbiol. 2020, 77, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Hou, Y.; Li, W.; Yang, S.; Li, Q.; Ziniu, Y. Biodiesel production from rice straw and restaurant waste employing black soldier fly assisted by microbes. Energy—Asia Pac. J. Energy Environ. 2012, 47, 225–229. [Google Scholar] [CrossRef]
- Xiao, X.; Mazza, L.; Yu, Y.; Cai, M.; Zheng, L.; Tomberlin, J.K.; Yu, J.; van Huis, A.; Yu, Z.; Fasulo, S.; et al. Efficient co-conversion process of chicken manure into protein feed and organic fertilizer by Hermetia illucens L. (Diptera: Stratiomyidae) larvae and functional bacteria. J. Environ. Manag. 2018, 217, 668–676. [Google Scholar] [CrossRef]
- Zheng, L.; Crippen, T.L.; Holmes, L.; Singh, B.; Pimsler, M.L.; Benbow, M.E.; Tarone, A.M.; Dowd, S.; Yu, Z.; Vanlaerhoven, S.L.; et al. Bacteria mediate oviposition by the black soldier fly, Hermetia illucens (L.), (Diptera: Stratiomyidae). Sci. Rep. 2013, 3, 2563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vikram, A.; Prithiviraj, B.; Hamzehzarghani, H.; Kushalappa, A. Volatile metabolite profiling to discriminate diseases of McIntosh apple inoculated with fungal pathogens. J. Sci. Food Agric. 2004, 84, 1333–1340. [Google Scholar] [CrossRef]
- Salvador, Â.; Baptista, I.; Barros, A.; Gomes, N.; Cunha, A.; Almeida, A.; Rocha, S. Can volatile organic metabolites be used to simultaneously assess microbial and mite contamination level in cereal grains and coffee beans? PLoS ONE 2013, 8, e59338. [Google Scholar]
- Lippolis, V.; Pascale, M.; Cervellieri, S.; Damascelli, A.; Visconti, A. Screening of deoxynivalenol contamination in durum wheat by MOS-based electronic nose and identification of the relevant pattern of volatile compounds. Food Control 2014, 37, 263–271. [Google Scholar] [CrossRef]
- Dahanukar, A.; Hallem, E.A.; Carlson, J.R. Insect chemoreception. Curr. Opin. Neurobiol. 2005, 15, 423–430. [Google Scholar] [CrossRef]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Pelosi, P.; Maida, R. Odorant-binding proteins in insects. Comp. Biochem. Physiol. B 1995, 111, 503–514. [Google Scholar] [CrossRef]
- Sun, Y.F.; De Biasio, F.; Qiao, H.L.; Iovinella, I.; Yang, S.X.; Ling, Y.; Riviello, L.; Battaglia, D.; Falabella, P.; Yang, X.L.; et al. Two odorant-binding proteins mediate the behavioural response of aphids to the alarm pheromone (E)-ß-farnesene and structural analogues. PLoS ONE 2012, 7, e32759. [Google Scholar]
- Vogt, R.G.; Große-Wilde, E.; Zhou, J.J. The Lepidoptera Odorant Binding Protein gene family: Gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 2015, 62, 142–153. [Google Scholar] [CrossRef]
- Venthur, H.; Zhou, J.J. Odorant Receptors and Odorant-Binding Proteins as Insect Pest Control Targets: A Comparative Analysis. Front. Physiol. 2018, 9, 1163. [Google Scholar] [CrossRef] [PubMed]
- Dennis, A.B.; Ballesteros, G.I.; Robin, S.; Schrader, L.; Bast, J.; Berghöfer, J.; Beukeboom, L.W.; Belghazi, M.; Bretaudeau, A.; Buellesbach, J.; et al. Functional insights from the GC-poor genomes of two aphid parasitoids, Aphidius ervi and Lysiphlebus fabarum. BMC Genom. 2020, 21, 376. [Google Scholar] [CrossRef] [PubMed]
- Vieira, F.G.; Rozas, J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 2011, 3, 476–490. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 2017, 93, 184–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelosi, P.; Calvello, M.; Ban, L. Diversity of Odorant-binding Proteins and Chemosensory Proteins in Insects. Chem. Senses 2005, 30 (Suppl. S1), i291–i292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.J.; Zhang, G.A.; Huang, W.; Birkett, M.A.; Field, L.M.; Pickett, J.A.; Pelosi, P. Revisiting the odorant binding protein LUSH of Drosophila melanogaster: Evidence for odour recognition and discrimination. FEBS Lett. 2004, 558, 23–26. [Google Scholar] [CrossRef] [Green Version]
- Hekmat-Scafe, D.S.; Scafe, C.R.; McKinney, A.J.; Tanouye, M.A. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002, 12, 1357–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstock, G.M.; Robinson, G.E.; Gibbs, R.A.; Worley, K.C.; Evans, J.D.; Maleszka, R.; Robertson, H.M.; Weaver, D.B.; Beye, M.; Bork, P.; et al. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931–949. [Google Scholar]
- Xu, P.X.; Zwiebel, L.J.; Smith, D.P. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 2003, 12, 549–560. [Google Scholar] [CrossRef]
- Hogsette, J.A. New diets for production of house flies and stable flies (Diptera: Muscidae) in the laboratory. J. Econ. Entomol. 1992, 85, 2291–2294. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Newell, P.D.; Fricker, A.D.; Roco, C.A.; Chandrangsu, P.; Merkel, S.M. A Small-Group Activity Introducing the Use and Interpretation of BLAST. J. Microbiol. Biol. Educ. 2013, 14, 238–243. [Google Scholar] [CrossRef] [Green Version]
- Kaleka, A.S.; Kaur, N.; Bali, G.K. Larval Development and Molting, Edible Insects, Heimo Mikkola, IntechOpen. 2019. Available online: https://www.intechopen.com/books/edible-insects/larval-development-and-molting (accessed on 20 February 2021).
- Georgescu, B.; Struti, D.; Păpuc, T.; Ladosi, D.; Boaru, A. Body weight loss of black soldier fly Hermetia illucens (Diptera: Stratiomyidae) during development in non-feeding stages: Implications for egg clutch parameters. Eur. J. Entomol. 2020, 117, 216–225. [Google Scholar] [CrossRef]
- JMP Pro, Version 15; 1989–2019; SAS Institute, Inc.: Cary, NC, USA.
- Zhang, Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins 2009, 77, 100–113. [Google Scholar] [CrossRef] [Green Version]
- DeLano, W.L.; Lam, J. PyMOL: A communications tool for computational models. Abstr. Pap. Am. Chem. Soc. 2005, 230, U1371–U1372. [Google Scholar]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [Green Version]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography ofproteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- Tomberlin, J.; Sheppard, D. Lekking Behavior of the Black Soldier Fly [Diptera: Stratiomyidae]. Fla. Entomol. 2001, 84, 729730. [Google Scholar] [CrossRef]
- Chia, S.; Tanga, C.; Khamis, F.; Mohamed, S.; Salifu, D.; Subramanian, S.; Fiaboe, K.; Niassy, S.; van Loon, J.; Dicke, M.; et al. Threshold temperatures and thermal requirements of black soldier fly Hermetia illucens: Implications for mass production. PLoS ONE 2018, 13, e0206097. [Google Scholar] [CrossRef] [Green Version]
- Sripontan, Y.; Juntavimon, T.; Chiu, I.C. Egg-trapping of black soldier fly, Hermetia illucens (L.) (Diptera: Stratiomyidae) with various wastes and the effects of environmental factors on egg-laying. Khon Kaen Agric. J. 2017, 45, 179–184. [Google Scholar]
- Nyakeri, E.M.; Ogola, H.J.O.; Amimo, F.A.; Ayieko, M.A. Comparison of the performance of different baiting attractants in the egg laying activity of the black soldier fly (Hermetia illucens L.). J. Entomol. Zool. Stud. 2017, 5, 153–158. [Google Scholar]
- Zheng, W.; Peng, W.; Zhu, C.; Zhang, Q.; Saccone, G.; Zhang, H. Identification and expression profile analysis of odorant binding proteins in the oriental fruit fly Bactrocera Dors. Int. J. Mol. Sci 2013, 14, 14936–14949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Coates, B.S.; Ge, X.; Bai, S.; He, K.; Wang, Z. Male- and Female-Biased Gene Expression of Olfactory-Related Genes in the Antennae of Asian Corn Borer, Ostrinia furnacalis (Guenée) (Lepidoptera: Crambidae). PLoS ONE 2015, 10, e0128550. [Google Scholar]
- Campanini, E.B.; Congrains, C.; Torres, F.R. Odorant-binding proteins expression patterns in recently diverged species of Anastrepha fruit flies. Sci. Rep. 2017, 7, 2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.Q.; Sun, H.Z.; Du, J. Identification and tissue distribution of chemosensory protein and odorant binding protein genes in Tropidothorax elegans Distant (Hemiptera: Lygaeidae). Sci. Rep. 2018, 8, 7803. [Google Scholar] [CrossRef] [PubMed]
- Ewusie, E.A.; Kwapong, P.K.; Ofosu-Budu, G.; Sandrock, C.; Akumah, A.M.; Nartey, E.K.; Tetegaga, C.; Agyakwah, S.K. The black soldier fly, Hermetia illucens (Diptera: Stratiomyidae): Trapping and culturing of wild colonies in Ghana. Sci. Afr. 2019, 5, e00134. [Google Scholar] [CrossRef]
- Bruno, D.; Grossi, G.; Salvia, R.; Scala, A.; Farina, D.; Grimaldi, A.; Zhou, J.-J.; Bufo, S.A.; Vogel, H.; Grosse-Wilde, E.; et al. Sensilla Morphology and Complex Expression Pattern of Odorant Binding Proteins in the Vetch Aphid Megoura viciae (Hemiptera: Aphididae). Front. Physiol. 2018, 9, 777. [Google Scholar] [CrossRef] [Green Version]
- De Biasio, F.; Riviello, L.; Bruno, D.; Grimaldi, A.; Congiu, T.; Sun, Y.F.; Falabella, P. Expression pattern analysis of odorant-binding proteins in the pea aphid Acyrthosiphon pisum. Insect Sci. 2015, 2, 220–234. [Google Scholar] [CrossRef]
- Gu, S.H.; Sun, L.; Yang, R.-N.; Wu, K.-M.; Guo, Y.-Y.; Li, X.-C.; Zhou, J.-J.; Zhang, Y.-J. Molecular characterization and differential expression of olfactory genes in the antennae of the black cutworm moth Agrotis ipsilon. PLoS ONE 2014, 9, e103420. [Google Scholar] [CrossRef] [Green Version]
- Calvello, M.; Guerra, N.; Brandazza, A.; D’Ambrosio, C.; Scaloni, A.; Dani, F.R.; Turillazzi, S.; Pelosi, P. Soluble proteins of chemical communication in the social wasp Polistes Dominulus. Cell Mol. Life Sci. 2003, 60, 1933–1943. [Google Scholar] [CrossRef]
- Zhou, C.X.; Min, S.F.; Tang, Y.L.; Wang, M.Q. Analysis of antennal transcriptome and odorant binding protein expression profiles of the recently identified parasitoid wasp, Sclerodermus sp. Comp. Biochem. Physiol. 2015, 16, 10–19. [Google Scholar] [CrossRef]
- Sun, J.S.; Xiao, S.; Carlson, J.R. The diverse small proteins called odorant-binding proteins. Open Biol. 2018, 8, 180208. [Google Scholar] [CrossRef] [Green Version]
- Harada, E.; Haba, D.; Aigaki, T.; Matsuo, T. Behavioral analyses of mutants for two odorant-binding protein genes, Obp57d and Obp57e, in Drosophila melanogaster. Genes Genet. Syst. 2008, 83, 257–264. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, T. Genes for host-plant selection in Drosophila. J. Neurogen. 2008, 22, 195–210. [Google Scholar] [CrossRef]
- Diehl, P.A.; Vlimant, M.; Guerenstein, P.; Guerin, P.M. Ultrastructure and receptor cell responses of the antennal grooved peg sensilla of Triatoma infestans (Hemiptera: Reduviidae). Arthropod Struct. Dev. 2003, 31, 271–285. [Google Scholar] [CrossRef] [Green Version]
- Pezzi, M.; Leis, M.; Chicca, M.; Falabella, P.; Salvia, R.; Scala, A.; Whitmore, D. Morphology of the Antenna of Hermetia illucens (Diptera: Stratiomyidae): An Ultrastructural Investigation. J. Med. Entomol. 2017, 54, 925–933. [Google Scholar] [CrossRef]
- Benoit, J.B.; Vigneron, A.; Broderick, N.A.; Wu, Y.; Sun, J.S.; Carlson, J.R.; Aksoy, S.; Weiss, B.L. Symbiont induced odorant binding proteins mediate insect host hematopoiesis. eLife 2017, 6, e19535. [Google Scholar] [CrossRef]
- Jacquin-Joly, E.; Vogt, R.G.; François, M.C.; Nagnan-Le Meillour, P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. Senses 2001, 26, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, S.; Zhang, K.; Ren, L.; Ali, A.; Zhang, Y.; Zhou, J.; Guo, Y. Odorant binding characteristics of three recombinant odorant binding proteins in Microplitis mediator (Hymenoptera: Braconidae). J. Chem. Ecol. 2014, 40, 541–548. [Google Scholar] [CrossRef]
- Li, S.; Picimbon, J.F.; Ji, S.; Kan, Y.; Chuanling, Q.; Zhou, J.J.; Pelosi, P. Multiple functions of an odorant binding protein in the mosquito Aedes aegypti. Biochem. Biophys. Res. Commun. 2008, 372, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Pitts, R.J. A blood-free protein meal supporting oogenesis in the Asian tiger mosquito, Aedes albopictus (Skuse). J. Insect Physiol. 2014, 64, 1–6. [Google Scholar] [CrossRef]
- Elhag, O.; Zhou, D.; Song, Q.; Soomro, A.A.; Cai, M.; Zheng, L.; Yu, Z.; Zhang, J. Screening, Expression, Purification and Functional Characterization of Novel Antimicrobial Peptide Genes from Hermetia illucens (L.). PLoS ONE 2017, 12, e0169582. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.; Müller, A.; Heckel, D.G.; Gutzeit, H.; Vilcinskas, A. Nutritional immunology: Diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev. Comp. Immunol. 2018, 78, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Moalemiyan, M.; Vikram, A.; Kushalappa, A.C. Detection and discrimination of two fungal diseases of mango (cv. Keitt) fruits based on volatile metabolite profiles using GC/MS. Postharvest Biol. Technol. 2007, 45, 117–125. [Google Scholar] [CrossRef]
- Azeem, M.; Rajarao, G.K.; Nordenhem, H.; Nordlander, G.; Borg-karlson, A.K. Penicillium expansum volatiles reduce pine weevil attraction to host plants. J. Chem. Ecol. 2013, 39, 120–128. [Google Scholar] [CrossRef] [Green Version]
- Ragaert, P.; Devlieghere, F.; Loos, S.; Dewulf, J.; Van Langenhove, H.; Debevere, J. Metabolite production of yeasts on a strawberry-agar during storage at 7 degrees C in air and low oxygen atmosphere. Food Microbiol. 2006, 23, 154–161. [Google Scholar] [CrossRef]
- Lalander, C.H.; Fidjeland, J.; Diener, S.; Eriksson, S.; Vinnerås, B. High waste-to-biomass conversion and efficient Salmonella spp. reduction using black soldier fly for waste recycling. Agron. Sustain. Dev. 2015, 35, 261–271. [Google Scholar] [CrossRef]
- Lemfack, M.C.; Gohlke, B.O.; Toguem, S.M.T.; Preissner, S.; Piechulla, B.; Preissner, R. mVOC 2.0: A database of microbial volatiles. Nucleic Acids Res. 2018, 46, D1261–D1265. [Google Scholar] [CrossRef] [Green Version]
- Davis, T.S.; Crippen, T.L.; Hofstetter, R.W.; Tomberlin, J.K. Microbial Volatile Emissions as Insect Semiochemicals. J. Chem. Ecol. 2013, 39, 840–859. [Google Scholar] [CrossRef]
- Schulz, S.; Dickschat, J.S. Bacterial volatiles: The smell of small organisms. Nat. Prod. Rep. 2007, 24, 814–842. [Google Scholar] [CrossRef] [PubMed]
- Effmert, U.; Kalderas, J.; Warnke, R.; Piechulla, B. Volatile Mediated Interactions Between Bacteria and Fungi in the Soil. J. Chem. Ecol. 2012, 38, 665–703. [Google Scholar] [CrossRef] [PubMed]
- Wierda, R.L.; Fletcher, G.; Xu, L.; Dufour, J.P. Analysis of volatile compounds as spoilage indicators in fresh king salmon (Oncorhynchus tshawytscha) during storage using SPME-GC-MS. J. Agric. Food Chem. 2006, 54, 8480–8490. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Khan, A.L.; Ali, L.; Khan, A.R.; Waqas, M.; Hussain, J.; Lee, I.J.; Shin, J.H. Benzaldehyde as an insecticidal, antimicrobial, and antioxidant compound produced by Photorhabdus temperata M1021. J. Microbiol. 2015, 53, 127–133. [Google Scholar] [CrossRef]
- Forbes, S.L.; Perrault, K.A. Decomposition Odour Profiling in the Air and Soil Surrounding Vertebrate Carrion. PLoS ONE 2014, 9, 12. [Google Scholar] [CrossRef] [Green Version]
- Mikš-Krajnik, M.; Yoon, Y.J.; Ukuku, D.O.; Yuk, H.G. Volatile chemical spoilage indexes of raw Atlantic salmon (Salmo salar) stored under aerobic condition in relation to microbiological and sensory shelf lives. Food Microbiol. 2016, 53, 182–191. [Google Scholar] [CrossRef]
- Paczkowski, S.; Schutz, S. Post-mortem volatiles of vertebrate tissue. App. Microbiol. Biotechnol. 2011, 91, 917–935. [Google Scholar] [CrossRef] [Green Version]
- Quinn, B.P.; Carlson, D.A.; Geden, C.J.; Bernier, U.R.; Booth, M.M.; Hogsette, J.A., Jr. Attractants for Insects Such as Flies. U.S. Patent No. 8,053,464, 8 November 2011. [Google Scholar]
- Balogun, A.O.; Sotoudehniakarani, F.; McDonald, A.G. Thermo-kinetic, spectroscopic study of brewer’s spent grains and characterisation of their pyrolysis products. J. Anal. Appl. Pyrolysis 2017, 127, 8–16. [Google Scholar] [CrossRef]
- Zhan, S.; Fang, G.; Cai, M.; Kou, Z.; Xu, J.; Cao, Y.; Bai, L.; Zhang, Y.; Jiang, Y.; Luo, X.; et al. Genomic landscape and genetic manipulation of the black soldier fly Hermetia illucens, a natural waste recycler. Cell Res. 2020, 30, 50–60. [Google Scholar] [CrossRef]
- Pelosi, P.; Zhu, J.; Knoll, W. Odorant-Binding Proteins as Sensing Elements for Odour Monitoring. Sensors 2018, 18, 3248. [Google Scholar] [CrossRef] [Green Version]



| Larval Contig vs. Adult Contig | Query Cover | e-Value | Identity |
|---|---|---|---|
| L21961 A41232 | 100% | 7 × 10−104 | 99.29% |
| L13368 A30834 | 100% | 1 × 10−104 | 100.00% |
| L57 A2226 | 100% | 4 × 10−103 | 100.00% |
| L11107 A11394 | 100% | 2 × 10−105 | 100.00% |
| L45961 A43028 | 100% | 8 × 10−134 | 100.00% |
| L768 A11523 | 100% | 4 × 10−100 | 98.55% |
| L1173 A8987 | 100% | 3 × 10−103 | 99.25% |
| L1844 A410 | 100% | 7 × 10−97 | 97.71% |
| L2633 A1002 | 100% | 2 × 10−94 | 96.15% |
| L2633 A1003 | 100% | 6 × 10−98 | 99.23% |
| L3948 A23172 | 100% | 8 × 10−102 | 99.26% |
| L3982 A2731 | 100% | 3 × 10−101 | 99.26% |
| L9011 A8085 | 100% | 7 × 10−105 | 100.00% |
| L13738 A3768 | 100% | 9 × 10−101 | 100.00% |
| L59460 A17957 | 100% | 2 × 10−108 | 95.45% |
| Time Point | Compound | Indicator Value | p Value | MVOC |
|---|---|---|---|---|
| Before BSF Feeding | n-propyl acetate | 0.9679 | 0.001 | Y |
| 2-methyl-butanal | 0.9304 | 0.001 | Y | |
| acetic acid, butyl ester | 0.9230 | 0.001 | Y | |
| butanoic acid, butyl ester | 0.8945 | 0.001 | Y | |
| acetic acid, hexyl ester | 0.8691 | 0.001 | Y | |
| 1-hexanol | 0.8518 | 0.001 | Y | |
| butyl 2-methylbutanoate | 0.8496 | 0.001 | Y | |
| acetic acid, pentyl ester | 0.8465 | 0.001 | Y | |
| beta pinene | 0.8417 | 0.001 | Y | |
| 2-pentanol, acetate | 0.8253 | 0.001 | N | |
| butanoic acid, propyl ester | 0.8122 | 0.001 | Y | |
| 2-hexenal | 0.8071 | 0.001 | Y | |
| acetic acid, 2-methylpropyl ester | 0.7692 | 0.001 | Y | |
| 2-hexen-1-ol, (E) | 0.7871 | 0.001 | Y | |
| propanoic acid, ethyl ester | 0.7423 | 0.001 | Y | |
| 1-butanol, 3-methyl-, acetate | 0.6411 | 0.001 | Y | |
| 2-hexen-1-ol, acetate, (E) | 0.6018 | 0.001 | N | |
| 3-methyl-butanal | 0.5989 | 0.009 | Y | |
| alpha pinene | 0.5852 | 0.049 | Y | |
| 1-butanol, 2-methyl-, acetate | 0.5109 | 0.001 | Y | |
| acetic acid, 1-methylethyl ester | 0.5000 | 0.001 | Y | |
| propanoic acid, butyl ester | 0.4867 | 0.001 | Y | |
| butanoic acid, 2-methyl-, ethyl ester | 0.4681 | 0.001 | Y | |
| hexanoic acid, ethyl ester | 0.4435 | 0.001 | Y | |
| hexanoic acid, hexyl ester | 0.4226 | 0.001 | N | |
| 3-methyl-2-butanol | 0.4126 | 0.001 | Y | |
| butanoic acid, 2-methyl-, propyl ester | 0.3902 | 0.001 | N | |
| butanoic acid, 2-methyl, hexyl ester | 0.3614 | 0.002 | N | |
| butanoic acid, 3-methylbutyl ester | 0.3284 | 0.046 | Y | |
| hexanoic acid, butyl ester | 0.3036 | 0.001 | Y | |
| butanoic acid, 1-methylethyl ester | 0.2917 | 0.001 | N | |
| propanoic acid, 1-methylethyl ester | 0.2500 | 0.002 | N | |
| propanoic acid, propyl ester | 0.1455 | 0.030 | Y | |
| After BSF Feeding | styrene | 0.9332 | 0.001 | Y |
| 4-methyl octane | 0.8898 | 0.001 | N | |
| acetophenone | 0.8898 | 0.001 | Y | |
| 2,4-dimethyl-1-heptene | 0.8554 | 0.001 | N | |
| 1,4-dichloro-benzene | 0.8179 | 0.001 | N | |
| alpha farnesene | 0.7572 | 0.001 | Y | |
| 4-methyl heptane | 0.7538 | 0.001 | Y | |
| 3-hydroxy-2-butanone | 0.7121 | 0.001 | Y | |
| delta limonene | 0.6809 | 0.001 | Y | |
| 3-octanone | 0.6349 | 0.009 | Y | |
| 2-hexanone | 0.3896 | 0.004 | Y |
| Diet Treatment | Compound | Indicator Value | p Value | MVOC |
|---|---|---|---|---|
| A | propanoic acid, propyl ester | 0.3411 | 0.001 | Y |
| hexanoic acid, butyl ester | 0.3295 | 0.002 | Y | |
| butanoic acid, 2-methyl-, propyl ester | 0.3082 | 0.005 | N | |
| hexanoic acid, hexyl ester | 0.3039 | 0.003 | N | |
| propanoic acid, butyl ester | 0.2970 | 0.002 | Y | |
| butanoic acid, 2-methyl, hexyl ester | 0.2919 | 0.003 | N | |
| alpha farnesene | 0.2772 | 0.020 | Y | |
| butanoic acid, propyl ester | 0.2767 | 0.015 | Y | |
| acetic acid, pentyl ester | 0.2683 | 0.009 | Y | |
| butanoic acid, 2-methyl-, ethyl ester | 0.2521 | 0.017 | Y | |
| butyl 2-methylbutanoate | 0.2521 | 0.024 | Y | |
| 1-butanol, 2-methyl-, acetate | 0.2416 | 0.018 | Y | |
| 2-hexen-1-ol, acetate, (E) | 0.2165 | 0.018 | N | |
| B | 2-pentanone | 0.2647 | 0.016 | Y |
| butanoic acid, 3-methyl-, 3-methylbutyl ester | 0.2385 | 0.035 | Y | |
| 3-methyl-2-butanol | 0.1969 | 0.030 | Y | |
| SG | acetic acid, 1-methylethyl ester | 0.2071 | 0.022 | Y |
| acetic acid, 1-methylpropyl ester | 0.2019 | 0.011 | Y | |
| propanoic acid, 1-methylethyl ester | 0.1630 | 0.040 | Y | |
| AB | 2-heptanone | 0.2574 | 0.032 | Y |
| Diet Treatment | Compound | t-Statistic | p Value | Response to BSF Feeding |
|---|---|---|---|---|
| A | acetic acid, 1-methylethyl ester | 2.0330 | 0.0307 | Decreased |
| 2-methyl-butanal | 7.6570 | <0.0001 | Decreased | |
| propanoic acid, ethyl ester | 6.1018 | <0.0001 | Decreased | |
| n-propyl acetate | 12.5324 | <0.0001 | Decreased | |
| 3-methyl 1 butanol | −2.6771 | 0.0090 | Increased | |
| 2-hexanone | −2.1064 | 0.0268 | Increased | |
| acetic acid, butyl ester | 9.0735 | <0.0001 | Decreased | |
| butanoic acid, 1-methylethyl ester | 3.2234 | 0.0031 | Decreased | |
| 2-hexen-1-ol, (E) | 13.3337 | <0.0001 | Decreased | |
| 1-hexanol | 17.6178 | <0.0001 | Decreased | |
| 2-heptanone | −3.4129 | 0.0021 | Increased | |
| styrene | −3.9625 | 0.0007 | Increased | |
| butanoic acid, propyl ester | 8.0026 | <0.0001 | Decreased | |
| 3-octanone | −2.6621 | 0.0093 | Increased | |
| hexanoic acid, ethyl ester | 3.6954 | 0.0012 | Decreased | |
| acetophenone | −2.9457 | 0.0053 | Increased | |
| butanoic acid, 2-methyl, hexyl ester | 6.8270 | <0.0001 | Decreased | |
| B | 2-methyl-butanal | 5.2414 | <0.0001 | Decreased |
| n-propyl acetate | 4.1729 | 0.0005 | Decreased | |
| acetic acid, 2-methylpropyl ester | 2.8090 | 0.007 | Decreased | |
| 2-hexanone | −2.7025 | 0.0086 | Increased | |
| acetic acid, butyl ester | 4.6626 | 0.0002 | Decreased | |
| 2-hexen-1-ol, (E) | 4.1726 | 0.0005 | Decreased | |
| 1-hexanol | 4.4551 | 0.0003 | Decreased | |
| 1-butanol, 3-methyl-, acetate | 2.8739 | 0.0061 | Decreased | |
| styrene | −3.0277 | 0.0045 | Increased | |
| 3-octanone | −3.0591 | 0.0042 | Increased | |
| delta limonene | −1.8663 | 0.0415 | Increased | |
| acetophenone | −2.9461 | 0.0053 | Increased | |
| alpha farnesene | −2.9470 | 0.0053 | Increased | |
| SG | acetic acid, 1-methylethyl ester | 3.1821 | 0.0033 | Decreased |
| 2-methyl-butanal | 1.7853 | 0.0479 | Decreased | |
| 2-pentanone | −2.6189 | 0.0101 | Increased | |
| propanoic acid, ethyl ester | 23.4326 | <0.0001 | Decreased | |
| n-propyl acetate | 5.6582 | <0.0001 | Decreased | |
| propanoic acid, 1-methylethyl ester | 2.6301 | 0.0099 | Decreased | |
| acetic acid, 2-methylpropyl ester | 3.9253 | 0.0008 | Decreased | |
| acetic acid, butyl ester | 3.7345 | 0.0011 | Decreased | |
| butanoic acid, 1-methylethyl ester | 2.5664 | 0.0112 | Decreased | |
| 1-hexanol | 3.1753 | 0.0034 | Decreased | |
| 1-butanol, 3-methyl-, acetate | 3.1919 | 0.0033 | Decreased | |
| styrene | −2.6347 | 0.0098 | Increased | |
| butanoic acid, propyl ester | 4.1825 | 0.0005 | Decreased | |
| delta limonene | −2.2986 | 0.0187 | Increased | |
| acetophenone | −2.7377 | 0.0080 | Increased | |
| alpha farnesene | −2.7992 | 0.0071 | Increased | |
| AB | 2-methyl-butanal | 4.2259 | 0.0004 | Decreased |
| n-propyl acetate | 5.4622 | <0.0001 | Decreased | |
| acetic acid, 2-methylpropyl ester | 3.3697 | 0.0023 | Decreased | |
| acetic acid, butyl ester | 5.5960 | <0.0001 | Decreased | |
| 2-hexen-1-ol, (E) | 5.3248 | <0.0001 | Decreased | |
| 1-hexanol | 3.3854 | 0.0022 | Decreased | |
| 1-butanol, 3-methyl-, acetate | 3.2337 | 0.0030 | Decreased | |
| styrene | −2.7290 | 0.0082 | Increased | |
| butanoic acid, propyl ester | 4.1006 | 0.0005 | Decreased | |
| 3-octanone | −1.8185 | 0.0452 | Increased | |
| acetophenone | −2.1429 | 0.0251 | Increased | |
| ASG | acetic acid, 1-methylethyl ester | 3.4404 | 0.0020 | Decreased |
| 2-methyl-butanal | 18.781 | <0.0001 | Decreased | |
| propanoic acid, ethyl ester | 24.1909 | <0.0001 | Decreased | |
| n-propyl acetate | 9.9488 | <0.0001 | Decreased | |
| 3-methyl 1 butanol | 3.1033 | 0.0039 | Decreased | |
| propanoic acid, 1-methylethyl ester | 2.5636 | 0.0113 | Decreased | |
| acetic acid, 2-methylpropyl ester | 7.5709 | <0.0001 | Decreased | |
| acetic acid, butyl ester | 18.0855 | <0.0001 | Decreased | |
| butanoic acid, 1-methylethyl ester | 2.3669 | 0.0164 | Decreased | |
| 2-hexen-1-ol, (E) | 3.9159 | 0.0008 | Decreased | |
| 1-hexanol | 6.0055 | <0.0001 | Decreased | |
| 1-butanol, 3-methyl-, acetate | 12.6400 | <0.0001 | Decreased | |
| styrene | −2.6991 | 0.0086 | Increased | |
| butanoic acid, propyl ester | 4.0969 | 0.0005 | Decreased | |
| hexanoic acid, ethyl ester | 7.4051 | <0.0001 | Decreased | |
| delta limonene | −1.8678 | 0.0414 | Increased | |
| butanoic acid, 2-methyl, hexyl ester | 4.0931 | 0.0005 | Decreased | |
| alpha farnesene | −1.8727 | 0.0411 | Increased | |
| BSG | 3-methyl butanal | 1.9782 | 0.0347 | Decreased |
| acetic acid, 1-methylethyl ester | 3.1037 | 0.0042 | Decreased | |
| 1-butanol | 1.8738 | 0.0418 | Decreased | |
| 2-methyl-butanal | 5.3343 | <0.0001 | Decreased | |
| 2-pentanone | 3.0636 | 0.0045 | Decreased | |
| propanoic acid, ethyl ester | 3.0325 | 0.0048 | Decreased | |
| n-propyl acetate | 6.3571 | <0.0001 | Decreased | |
| 3-methyl 1 butanol | 2.4682 | 0.0141 | Decreased | |
| propanoic acid, 1-methylethyl ester | 2.3981 | 0.0161 | Decreased | |
| acetic acid, 2-methylpropyl ester | 5.5777 | <0.0001 | Decreased | |
| acetic acid, butyl ester | 14.2486 | <0.0001 | Decreased | |
| 2-hexen-1-ol, (E) | 5.2596 | <0.0001 | Decreased | |
| 1-hexanol | 9.2635 | <0.0001 | Decreased | |
| 1-butanol, 3-methyl-, acetate | 6.0414 | <0.0001 | Decreased | |
| styrene | −2.4923 | 0.0135 | Increased | |
| butanoic acid, propyl ester | 2.5515 | 0.0121 | Decreased | |
| acetophenone | −1.7904 | 0.0483 | Increased | |
| alpha farnesene | −2.4558 | 0.0144 | Increased |
| HillOBPs 3D Structure | Highlighted Residues Involved in Ligand Binding | Number of Pockets | Mouths | Area of Solvent Access Surface (Å2) | Area of Molecular Surface (Å2) | Volume of Solvent Access Surface (Å3) | Volume of Molecular Surface (Å3) | Diameter Pocket (Å) | Hydro-Phobicity (%) |
|---|---|---|---|---|---|---|---|---|---|
 | >HillOBP_C57 tytikthddliktrglcvkelnvpdnyvekfkkwdfqddettrcyikcvlnkmelfdtangfnvenlveqlgqnkdktevrtevtkcsdkneqksddctwayrgfkcflskhlqlvqssvks | 16 | 1 | 275.84 | 615.78 | 116.07 | 729.82 | 285.99 | 79.31 |
 | >HillOBP_C11107 ewvprtsdqmykdqaecfkqlelteeeqqkvkkedfpdepkfrcylrcilmggqiwddekgynperayaellnidmtadvenlrkcntqnlhhsdsctrafrvvkcfannnyitsikpks | 21 | 4 | 338.23 | 753.68 | 185.62 | 901.17 | 316.48 | 56.67 |
 | >HillOBP_C21691 nvndpklksileqcigsekaspadiaalearssdlskeakcviscvmknykllsddgkvnrdvfmaeaeemtkgdagamkeagemfeicsaktvadpcesafnfghcmktemtarnipmdf | 17 | 0 | 228.54 | 579.12 | 83.35 | 628.65 | 258.62 | 72.41 |
 | >HillOBP_C1173 nwstptkeqfkqhrddclkegnvpeetankirkeqypndrdtycyircvgsksgiwndrkgydidrslqvfeangyevtrenlercfaplpgadtctwagvnmrclrdnkyvtkkasa | 19 | 1 | 313.85 | 807.34 | 136.89 | 900.72 | 378.30 | 53.33 |
 | >HillOBP_C2633 isteefqemreecfksekvpeadieklkhreygldlgheakcyirclgmktgnwddtngydvekiytdfrtaglevtkenlkkcfkssgdddkcvwaakdlkclwtnkymsrkq | 15 | 2 | 382.45 | 654.31 | 285.81 | 1005.38 | 334.98 | 53.85 |
 | >HillOBP_C13368 dvndprlkaslekcigsekaspadvealkahssdlsreaqcvmacvmkefkllgddgkinrdvymaeaeemakgdagaikqatemydicsaktvadncesannfgqciknemiarnipldm | 15 | 1 | 379.24 | 841.48 | 199.76 | 1011.53 | 376.49 | 69.70 |
 | >HillOBP_C13738 dwkprsreqytkdgdecfksenisedgiheirrhvftddskcffrcvlmknhvwddttgynvervykevthiglkaskdgltqcnsddkkdkdpcqwvnnivrcvfehnyiepny | 20 | 2 | 240.94 | 454.29 | 83.13 | 534.66 | 227.29 | 64.71 |
 | >HillOBP_C31956 kvdenklkaytaniaktcqpegepfgevhdivekanptqdekcfitctmtkwgllsengkfqpdgvrkvneairefddnpaeyknadeaiiakcsaiekpekcdkgyaiaecgfkvfdeihg | 17 | 1 | 493.53 | 802.17 | 398.72 | 1286.26 | 360.19 | 62.16 |
| Energy (ΔG, kcal/mol) | ||||||||
|---|---|---|---|---|---|---|---|---|
| VOCs | OBP_C57 | OBP_C1173 | OBP_C2633 | OBP_C13368 | OBP_C31956 | OBP_C13738 | OBP_C21691 | OBP_C11107 |
| hexanoic acid, hexyl ester | −47.94 | −44.71 | −47.88 | −50.04 | −42.63 | −44.10 | −44.80 | −46.54 |
| hexanoic acid, butyl ester | −43.91 | −38.89 | −41.47 | −43.30 | −38.31 | −36.80 | −46.54 | −40.07 |
| isopentyl hexanoate | −41.45 | −38.45 | −42.33 | −43.53 | −37.59 | −38.02 | −40.16 | −39.96 |
| butanoic acid, 2-methyl, hexyl ester | −41.05 | −37.01 | −41.14 | −44.11 | −38.98 | −34.96 | −39.68 | −39.93 |
| butanoic acid, butyl ester | −37.05 | −32.86 | −34.51 | −36.28 | −33.00 | −32.86 | −36.49 | −34.70 |
| acetic acid, hexyl ester | −36.18 | −32.31 | −35.05 | −33.57 | −30.26 | −29.41 | −33.80 | −32.42 |
| 4-methyl octane | −35.89 | −32.32 | −34.26 | −36.00 | −30.94 | −28.77 | −32.71 | −32.81 |
| alpha-farnesene | −35.28 | −24.42 | −28.61 | −32.20 | −24.74 | −23.14 | −23.83 | −28.01 |
| butyl 2-methylbutanoate | −35.25 | −32.64 | −33.22 | −37.15 | −32.51 | −32.64 | −36.70 | −34.74 |
| hexanoic acid, ethyl ester | −35.04 | −33.63 | −33.06 | −36.18 | −32.34 | −32.72 | −37.84 | −33.35 |
| butanoic acid, 3-methylbutyl ester | −34.38 | −33.09 | −34.26 | −36.28 | −31.53 | −31.87 | −34.61 | −32.56 |
| butanoic acid, 1-methylbutyl ester | −34.38 | −32.49 | −35.52 | −35.03 | −30.97 | −32.83 | −34.21 | −33.22 |
| butanoic acid, 3-methyl-3-methylbutyl ester | −33.76 | −30.52 | −34.29 | −36.00 | −29.83 | −32.11 | −33.62 | −32.76 |
| 3-octanone | −33.71 | −31.42 | −35.07 | −33.79 | −30.43 | −30.12 | −33.11 | −31.79 |
| 2-hexen-1-ol, acetate, (E) | −33.38 | −29.82 | −33.25 | −33.62 | −28.98 | −30.13 | −28.31 | −31.81 |
| propanoic acid, butyl ester | −32.72 | −29.33 | −31.89 | −31.91 | −28.57 | −30.43 | −31.95 | −29.91 |
| 2-pentyl furan | −32.43 | −29.68 | −30.36 | −32.04 | −29.62 | −28.22 | −32.03 | −30.30 |
| butanoic acid, 2-methylpropyl ester | −31.90 | −29.04 | −31.43 | −33.34 | −29.51 | −29.81 | −33.93 | −30.38 |
| butanoic acid, propyl ester | −31.55 | −29.93 | −31.18 | −33.48 | −30.91 | −29.69 | −32.69 | −29.58 |
| 4-methyl heptane | −30.76 | −29.85 | −30.33 | −32.79 | −29.23 | −25.92 | −28.15 | −29.10 |
| acetic acid, pentyl ester | −30.72 | −27.43 | −30.94 | −31.29 | −28.75 | −28.36 | −31.82 | −30.26 |
| 2-heptanone | −29.46 | −26.79 | −29.01 | −29.35 | −25.71 | −26.06 | −29.62 | −27.76 |
| 1-hexanol | −29.32 | −25.85 | −25.81 | −29.88 | −26.30 | −26.15 | −30.47 | −26.77 |
| propanoic acid, propyl ester | −28.88 | −25.80 | −28.11 | −28.36 | −26.91 | −26.89 | −29.73 | −26.57 |
| butanoic acid, 1-methylethyl ester | −28.83 | −27.45 | −27.90 | −30.74 | −27.90 | −26.73 | −28.66 | −26.76 |
| butanoic acid, 2-methylethyl ester | −28.65 | −25.84 | −27.18 | −28.34 | −25.48 | −25.62 | −27.01 | −26.02 |
| 2-pentanol, acetate | −28.00 | −24.31 | −26.44 | −26.66 | −24.66 | −24.22 | −29.30 | −26.61 |
| delta-limonene | −27.91 | −26.76 | −28.21 | −29.21 | −26.70 | −22.69 | −21.09 | −27.35 |
| 2,4-dimethyl-1-heptene | −27.39 | −26.02 | −25.72 | −29.29 | −24.97 | −22.14 | −28.62 | −26.47 |
| 1-butanol, 2-methylacetate | −27.09 | −25.47 | −25.78 | −26.93 | −24.25 | −24.79 | −25.50 | −25.00 |
| acetic acid, butyl ester | −26.80 | −25.56 | −26.00 | −28.94 | −25.32 | −26.61 | −29.31 | −25.27 |
| 2-hexen-1-ol, (E) | −26.20 | −24.79 | −25.66 | −26.60 | −25.26 | −25.42 | −32.58 | −26.75 |
| 1-butanol, 3-methylacetate | −26.14 | −24.49 | −25.64 | −27.37 | −25.89 | −23.93 | −27.22 | −24.16 |
| 2-hexanone | −25.85 | −23.64 | −25.61 | −26.60 | −23.43 | −22.82 | −26.35 | −23.95 |
| propanoic acid, 1-methylethyl ester | −25.53 | −23.77 | −25.29 | −25.81 | −22.65 | −24.15 | −25.12 | −24.08 |
| n-propyl acetate | −24.80 | −21.68 | −22.74 | −26.14 | −21.99 | −22.52 | −25.69 | −22.64 |
| propanoic acid, ethyl ester | −24.69 | −22.47 | −24.29 | −25.05 | −22.39 | −24.17 | −26.50 | −23.16 |
| 2-hexenal | −23.33 | −22.47 | −22.57 | −23.08 | −21.96 | −21.73 | −26.85 | −22.35 |
| acetic acid, 1-methylpropyl ester | −23.29 | −22.29 | −23.13 | −24.17 | −21.49 | −20.94 | −24.45 | −22.29 |
| acetic acid, 2-methylpropyl ester | −22.76 | −22.32 | −23.70 | −25.17 | −21.75 | −22.03 | −23.71 | −22.07 |
| 2-pentanone | −21.72 | −19.76 | −22.39 | −21.59 | −19.42 | −19.93 | −23.73 | −20.49 |
| acetic acid, 1-methylethyl ester | −21.70 | −18.68 | −19.94 | −20.53 | −18.17 | −18.67 | −23.24 | −19.03 |
| 3-methyl butanal | −20.75 | −19.37 | −21.15 | −20.96 | −18.97 | −18.32 | −22.25 | −19.28 |
| 1-butanol | −20.60 | −20.04 | −20.22 | −21.18 | −18.71 | −20.06 | −22.19 | −19.72 |
| 2-methyl-1-butanol | −20.50 | −18.55 | −20.78 | −21.18 | −18.14 | −17.66 | −21.53 | −18.66 |
| 3-methyl-1-butanol | −20.32 | −19.39 | −21.77 | −20.13 | −18.91 | −17.74 | −20.15 | −18.92 |
| 2-methyl butanal | −19.51 | −19.58 | −20.86 | −20.94 | −17.96 | −18.02 | −22.68 | −19.45 |
| 3-methyl-2-butanol | −16.91 | −15.98 | −19.07 | −17.48 | −15.41 | −14.69 | −17.10 | −15.69 |
| 3-hydroxy-2-butanone | −14.95 | −14.92 | −16.23 | −16.18 | −13.73 | −14.84 | −16.17 | −14.14 |
| 1,4-dichlorobenzene | −7.00 | −4.91 | −6.12 | −6.56 | −5.80 | −5.84 | −8.19 | −5.46 |
| benzaldehyde | −2.42 | −2.43 | −3.83 | −2.84 | −1.82 | −2.91 | −5.34 | −2.08 |
| acetophenone | −2.38 | −1.47 | −3.29 | −2.60 | −0.95 | −2.57 | −4.99 | −2.91 |
| styrene | −1.18 | 0.42 | −0.49 | 0.69 | 0.40 | −0.55 | −3.34 | −0.33 |
| beta-pinene | 46.13 | 41.79 | 46.36 | 41.49 | 43.11 | 49.55 | 50.76 | 43.80 |
| alpha-pinene | 51.27 | 45.95 | 50.38 | 46.01 | 48.30 | 52.29 | 54.58 | 48.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scieuzo, C.; Nardiello, M.; Farina, D.; Scala, A.; Cammack, J.A.; Tomberlin, J.K.; Vogel, H.; Salvia, R.; Persaud, K.; Falabella, P. Hermetia illucens (L.) (Diptera: Stratiomyidae) Odorant Binding Proteins and Their Interactions with Selected Volatile Organic Compounds: An In Silico Approach. Insects 2021, 12, 814. https://0-doi-org.brum.beds.ac.uk/10.3390/insects12090814
Scieuzo C, Nardiello M, Farina D, Scala A, Cammack JA, Tomberlin JK, Vogel H, Salvia R, Persaud K, Falabella P. Hermetia illucens (L.) (Diptera: Stratiomyidae) Odorant Binding Proteins and Their Interactions with Selected Volatile Organic Compounds: An In Silico Approach. Insects. 2021; 12(9):814. https://0-doi-org.brum.beds.ac.uk/10.3390/insects12090814
Chicago/Turabian StyleScieuzo, Carmen, Marisa Nardiello, Donatella Farina, Andrea Scala, Jonathan A. Cammack, Jeffery K. Tomberlin, Heiko Vogel, Rosanna Salvia, Krishna Persaud, and Patrizia Falabella. 2021. "Hermetia illucens (L.) (Diptera: Stratiomyidae) Odorant Binding Proteins and Their Interactions with Selected Volatile Organic Compounds: An In Silico Approach" Insects 12, no. 9: 814. https://0-doi-org.brum.beds.ac.uk/10.3390/insects12090814