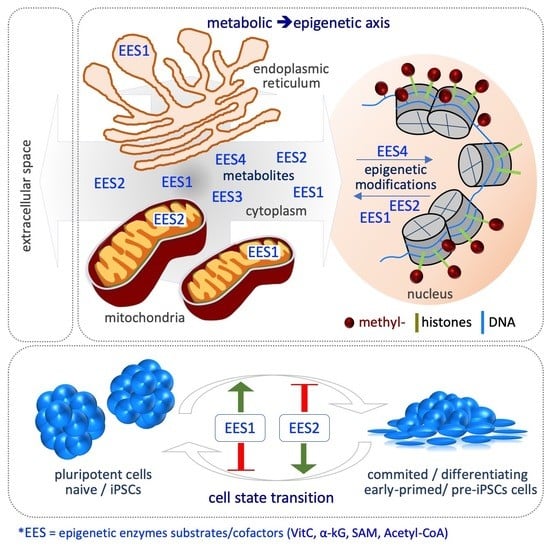
Metabolic–Epigenetic Axis in Pluripotent State Transitions
Abstract
:1. Introduction
2. Acetyl Coenzyme A
3. Nicotinamide Adenine Dinucleotide
4. Folate Coenzyme
5. Amino Acids
5.1. Threonine
5.2. Methionine/S-Adenosylmethionine
5.3. Proline
6. Alpha-Ketoglutarate
7. Vitamin C
8. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Tatapudy, S.; Aloisio, F.; Barber, D.; Nystul, T. Cell fate decisions: Emerging roles for metabolic signals and cell morphology. EMBO Rep. 2017, 18, 2105–2118. [Google Scholar] [CrossRef] [PubMed]
- Reid, M.A.; Dai, Z.; Locasale, J.W. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 2017, 19, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Choi, M.; Margineantu, D.; Margaretha, L.; Hesson, J.; Cavanaugh, C.; Blau, C.A.; Horwitz, M.S.; Hockenbery, D.; Ware, C.; et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012, 31, 2103–2116. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Gaeta, X.; Sahakyan, A.; Chan, A.B.; Hong, C.S.; Kim, R.; Braas, D.; Plath, K.; Lowry, W.E.; Christofk, H.R. Glycolytic Metabolism Plays a Functional Role in Regulating Human Pluripotent Stem Cell State. Cell Stem Cell 2016, 19, 476–490. [Google Scholar] [CrossRef] [Green Version]
- Varum, S.; Rodrigues, A.S.; Moura, M.B.; Momcilovic, O.; Easley, C.A.t.; Ramalho-Santos, J.; Van Houten, B.; Schatten, G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One 2011, 6, e20914. [Google Scholar] [CrossRef]
- Cliff, T.S.; Wu, T.; Boward, B.R.; Yin, A.; Yin, H.; Glushka, J.N.; Prestegaard, J.H.; Dalton, S. MYC Controls Human Pluripotent Stem Cell Fate Decisions through Regulation of Metabolic Flux. Cell Stem Cell 2017, 21, 502–516 e509. [Google Scholar] [CrossRef]
- Folmes, C.D.; Nelson, T.J.; Martinez-Fernandez, A.; Arrell, D.K.; Lindor, J.Z.; Dzeja, P.P.; Ikeda, Y.; Perez-Terzic, C.; Terzic, A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011, 14, 264–271. [Google Scholar] [CrossRef]
- Panopoulos, A.D.; Yanes, O.; Ruiz, S.; Kida, Y.S.; Diep, D.; Tautenhahn, R.; Herrerias, A.; Batchelder, E.M.; Plongthongkum, N.; Lutz, M.; et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012, 22, 168–177. [Google Scholar] [CrossRef]
- Zhang, J.; Nuebel, E.; Daley, G.Q.; Koehler, C.M.; Teitell, M.A. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 2012, 11, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, S.; Zhang, J.; Sun, Z.; Zhang, L.; Ross, C.A.; Huang, Y.C.; Asara, J.M.; Li, H.; Daley, G.Q.; Collins, J.J. Comprehensive Mapping of Pluripotent Stem Cell Metabolism Using Dynamic Genome-Scale Network Modeling. Cell Rep. 2017, 21, 2965–2977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryall, J.G.; Cliff, T.; Dalton, S.; Sartorelli, V. Metabolic Reprogramming of Stem Cell Epigenetics. Cell Stem Cell 2015, 17, 651–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivanand, S.; Viney, I.; Wellen, K.E. Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends Biochem. Sci. 2018, 43, 61–74. [Google Scholar] [CrossRef]
- Ye, C.; Tu, B.P. Sink into the Epigenome: Histones as Repositories That Influence Cellular Metabolism. Trends Endocrinol. Metab. 2018, 29, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Sutter, B.M.; Li, B.; Tu, B.P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 2011, 42, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Yanes, O.; Clark, J.; Wong, D.M.; Patti, G.J.; Sanchez-Ruiz, A.; Benton, H.P.; Trauger, S.A.; Desponts, C.; Ding, S.; Siuzdak, G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 2010, 6, 411–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Zhang, T.; Wang, L.; Cai, Y.; Zhong, X.; He, X.; Hu, L.; Tian, S.; Wu, M.; Hui, L.; et al. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 2017, 36, 1330–1347. [Google Scholar] [CrossRef] [Green Version]
- Shyh-Chang, N.; Daley, G.Q. Metabolic switches linked to pluripotency and embryonic stem cell differentiation. Cell Metab. 2015, 21, 349–350. [Google Scholar] [CrossRef]
- Moussaieff, A.; Rouleau, M.; Kitsberg, D.; Cohen, M.; Levy, G.; Barasch, D.; Nemirovski, A.; Shen-Orr, S.; Laevsky, I.; Amit, M.; et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015, 21, 392–402. [Google Scholar] [CrossRef]
- Miller, S.J. Cellular and physiological effects of short-chain fatty acids. Mini Rev. Med. Chem 2004, 4, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Chou, B.K.; Yen, J.; Ye, Z.; Zou, J.; Dowey, S.; Brodsky, R.A.; Ohm, J.E.; Yu, W.; Baylin, S.B.; et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 2010, 28, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Shyh-Chang, N.; Locasale, J.W.; Lyssiotis, C.A.; Zheng, Y.; Teo, R.Y.; Ratanasirintrawoot, S.; Zhang, J.; Onder, T.; Unternaehrer, J.J.; Zhu, H.; et al. Influence of Threonine Metabolism on S-Adenosylmethionine and Histone Methylation. Science 2012, 339, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Tang, S.; Li, X. Sirtuins in Metabolic and Epigenetic Regulation of Stem Cells. Trends Endocrinol. Metab. 2019, 30, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Calvanese, V.; Lara, E.; Suarez-Alvarez, B.; Abu Dawud, R.; Vazquez-Chantada, M.; Martinez-Chantar, M.L.; Embade, N.; Lopez-Nieva, P.; Horrillo, A.; Hmadcha, A.; et al. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 13736–13741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.; Huang, G.; Fan, W.; Chen, Y.; Ward, J.M.; Xu, X.; Xu, Q.; Kang, A.; McBurney, M.W.; Fargo, D.C.; et al. SIRT1-mediated deacetylation of CRABPII regulates cellular retinoic acid signaling and modulates embryonic stem cell differentiation. Mol Cell 2014, 55, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.O.; Taylor, A.K.; Bell, E.L.; Lim, R.; Kim, D.M.; Guarente, L. Sirtuin 1 Promotes Deacetylation of Oct4 and Maintenance of Naive Pluripotency. Cell Rep. 2016, 17, 809–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heo, J.; Lim, J.; Lee, S.; Jeong, J.; Kang, H.; Kim, Y.; Kang, J.W.; Yu, H.Y.; Jeong, E.M.; Kim, K.; et al. Sirt1 Regulates DNA Methylation and Differentiation Potential of Embryonic Stem Cells by Antagonizing Dnmt3l. Cell Rep. 2017, 18, 1930–1945. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Wang, T.T.; Liu, X.Z.; Wang, N.Y.; Sun, L.H.; Zhang, Z.Q.; Chen, H.Z.; Lv, X.; Huang, Y.; Liu, D.P. Sirt6 regulates efficiency of mouse somatic reprogramming and maintenance of pluripotency. Stem Cell Res. 2019, 10, 9. [Google Scholar] [CrossRef]
- Etchegaray, J.P.; Chavez, L.; Huang, Y.; Ross, K.N.; Choi, J.; Martinez-Pastor, B.; Walsh, R.M.; Sommer, C.A.; Lienhard, M.; Gladden, A.; et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat. Cell Biol. 2015, 17, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Son, M.J.; Son, M.Y.; Seol, B.; Kim, M.J.; Yoo, C.H.; Han, M.K.; Cho, Y.S. Nicotinamide overcomes pluripotency deficits and reprogramming barriers. Stem Cells 2013, 31, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Ren, Z.; Xu, F.; Zhou, X.; Song, C.; Wang, V.Y.; Liu, W.; Lu, L.; Thomson, J.A.; Chen, G. Nicotinamide Promotes Cell Survival and Differentiation as Kinase Inhibitor in Human Pluripotent Stem Cells. Stem Cell Rep. 2018, 11, 1347–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kropp, E.M.; Oleson, B.J.; Broniowska, K.A.; Bhattacharya, S.; Chadwick, A.C.; Diers, A.R.; Hu, Q.; Sahoo, D.; Hogg, N.; Boheler, K.R.; et al. Inhibition of an NAD(+) salvage pathway provides efficient and selective toxicity to human pluripotent stem cells. Stem Cells Transl. Med. 2015, 4, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Griffin, S.M.; Pickard, M.R.; Orme, R.P.; Hawkins, C.P.; Williams, A.C.; Fricker, R.A. Nicotinamide alone accelerates the conversion of mouse embryonic stem cells into mature neuronal populations. PLoS One 2017, 12, e0183358. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, D.E.; Pennington, B.O.; Croze, R.H.; Hinman, C.R.; Coffey, P.J.; Clegg, D.O. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl. Med. 2013, 2, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Nostro, M.C.; Sarangi, F.; Yang, C.; Holland, A.; Elefanty, A.G.; Stanley, E.G.; Greiner, D.L.; Keller, G. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Rep. 2015, 4, 591–604. [Google Scholar] [CrossRef]
- Idelson, M.; Alper, R.; Obolensky, A.; Ben-Shushan, E.; Hemo, I.; Yachimovich-Cohen, N.; Khaner, H.; Smith, Y.; Wiser, O.; Gropp, M.; et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell 2009, 5, 396–408. [Google Scholar] [CrossRef]
- Parsons, X.H.; Teng, Y.D.; Parsons, J.F.; Snyder, E.Y.; Smotrich, D.B.; Moore, D.A. Efficient derivation of human cardiac precursors and cardiomyocytes from pluripotent human embryonic stem cells with small molecule induction. J. Vis. Exp 2011, 57, e3274. [Google Scholar] [CrossRef]
- Vaca, P.; Berna, G.; Araujo, R.; Carneiro, E.M.; Bedoya, F.J.; Soria, B.; Martin, F. Nicotinamide induces differentiation of embryonic stem cells into insulin-secreting cells. Exp. Cell Res. 2008, 314, 969–974. [Google Scholar] [CrossRef]
- Mentch, S.J.; Locasale, J.W. One-carbon metabolism and epigenetics: Understanding the specificity. Ann. N. Y. Acad. Sci. 2016, 1363, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Xu, J.; Hu, W.; Li, Z.; Wu, J.; Zhang, S. Transient folate deprivation facilitates the generation of mouse-induced pluripotent stem cells. Cell Biol. Int. 2014, 38, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.T.; Yan, Q.Y.; Fang, Y.; Qiu, Z.D.; Zhang, S.M. Transient folate deprivation in combination with small-molecule compounds facilitates the generation of somatic cell-derived pluripotent stem cells in mice. J. Huazhong Univ. Sci. Technol. Med. Sci 2014, 34, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Kasulanati, S.; Venkatesan, V. Understanding pluripotency under folic acid deficiency using embryonic stem cells as an in vitro model. Med. Hypotheses 2018, 111, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Wang, L.; Guan, Y.; Shangguan, S.; Du, Q.; Wang, Y.; Zhang, T.; Zhang, Y. Long interspersed nucleotide element-1 hypomethylation in folate-deficient mouse embryonic stem cells. J. Cell Biochem. 2013, 114, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Jia, W.; Qian, Z.; Zhao, L.; Yu, Y.; Li, L.; Wang, C.; Zhang, W.; Liu, Q.; Yang, D.; et al. Folic Acid Supports Pluripotency and Reprogramming by Regulating LIF/STAT3 and MAPK/ERK Signaling. Stem Cells Dev. 2017, 26, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Li, Y.; Li, Z.; Liu, Z.; Zhang, Z.; Chang, S.; Wu, J. Mechanism of folate deficiency-induced apoptosis in mouse embryonic stem cells: Cell cycle arrest/apoptosis in G1/G0 mediated by microRNA-302a and tumor suppressor gene Lats2. Int. J. Biochem Cell Biol. 2012, 44, 1750–1760. [Google Scholar] [CrossRef] [PubMed]
- Fawal, M.A.; Jungas, T.; Kischel, A.; Audouard, C.; Iacovoni, J.S.; Davy, A. Cross Talk between One-Carbon Metabolism, Eph Signaling, and Histone Methylation Promotes Neural Stem Cell Differentiation. Cell Rep. 2018, 23, 2864–2873 e2867. [Google Scholar] [CrossRef] [PubMed]
- Valensisi, C.; Andrus, C.; Buckberry, S.; Doni Jayavelu, N.; Lund, R.J.; Lister, R.; Hawkins, R.D. Epigenomic Landscapes of hESC-Derived Neural Rosettes: Modeling Neural Tube Formation and Diseases. Cell Rep. 2017, 20, 1448–1462. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wang, Z.; Xie, Y.; Guo, X.; Tang, X.; Wang, S.; Yang, S.; Chen, K.; Niu, Y.; Ji, W. Folic acid deficiency inhibits neural rosette formation and neuronal differentiation from rhesus monkey embryonic stem cells. J. Neurosci. Res. 2012, 90, 1382–1391. [Google Scholar] [CrossRef]
- Sahakyan, V.; Duelen, R.; Tam, W.L.; Roberts, S.J.; Grosemans, H.; Berckmans, P.; Ceccarelli, G.; Pelizzo, G.; Broccoli, V.; Deprest, J.; et al. Folic Acid Exposure Rescues Spina Bifida Aperta Phenotypes in Human Induced Pluripotent Stem Cell Model. Sci. Rep. 2018, 8, 2942. [Google Scholar] [CrossRef] [PubMed]
- Kilberg, M.S.; Terada, N.; Shan, J. Influence of Amino Acid Metabolism on Embryonic Stem Cell Function and Differentiation. Adv. Nutr. 2016, 7, 780S–789S. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Alexander, P.; Wu, L.; Hammer, R.; Cleaver, O.; McKnight, S.L. Dependence of mouse embryonic stem cells on threonine catabolism. Science 2009, 325, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Alexander, P.B.; Wang, J.; McKnight, S.L. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc. Natl. Acad. Sci. USA 2011, 108, 15828–15833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, C.; Gu, H.; Wang, J.; Lu, W.; Mei, Y.; Wu, M. Regulation of L-threonine dehydrogenase in somatic cell reprogramming. Stem Cells 2013, 31, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, J. A regulatory circuitry locking pluripotent stemness to embryonic stem cell: Interaction between threonine catabolism and histone methylation. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef]
- Shiraki, N.; Shiraki, Y.; Tsuyama, T.; Obata, F.; Miura, M.; Nagae, G.; Aburatani, H.; Kume, K.; Endo, F.; Kume, S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014, 19, 780–794. [Google Scholar] [CrossRef]
- Fernandez-Arroyo, S.; Cuyas, E.; Bosch-Barrera, J.; Alarcon, T.; Joven, J.; Menendez, J.A. Activation of the methylation cycle in cells reprogrammed into a stem cell-like state. Oncoscience 2015, 2, 958–967. [Google Scholar]
- Tang, S.; Fang, Y.; Huang, G.; Xu, X.; Padilla-Banks, E.; Fan, W.; Xu, Q.; Sanderson, S.M.; Foley, J.F.; Dowdy, S.; et al. Methionine metabolism is essential for SIRT1-regulated mouse embryonic stem cell maintenance and embryonic development. EMBO J. 2017, 36, 3175–3193. [Google Scholar] [CrossRef]
- Chan, Y.S.; Goke, J.; Ng, J.H.; Lu, X.; Gonzales, K.A.; Tan, C.P.; Tng, W.Q.; Hong, Z.Z.; Lim, Y.S.; Ng, H.H. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 2013, 13, 663–675. [Google Scholar] [CrossRef]
- Duggal, G.; Warrier, S.; Ghimire, S.; Broekaert, D.; Van der Jeught, M.; Lierman, S.; Deroo, T.; Peelman, L.; Van Soom, A.; Cornelissen, R.; et al. Alternative Routes to Induce Naive Pluripotency in Human Embryonic Stem Cells. Stem Cells 2015, 33, 2686–2698. [Google Scholar] [CrossRef] [PubMed]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Ware, C.B.; Nelson, A.M.; Mecham, B.; Hesson, J.; Zhou, W.; Jonlin, E.C.; Jimenez-Caliani, A.J.; Deng, X.; Cavanaugh, C.; Cook, S.; et al. Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 4484–4489. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 2014, 158, 1254–1269. [Google Scholar] [CrossRef] [PubMed]
- Sperber, H.; Mathieu, J.; Wang, Y.; Ferreccio, A.; Hesson, J.; Xu, Z.; Fischer, K.A.; Devi, A.; Detraux, D.; Gu, H.; et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015, 17, 1523–1535. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Comes, S.; Gagliardi, M.; Laprano, N.; Fico, A.; Cimmino, A.; Palamidessi, A.; De Cesare, D.; De Falco, S.; Angelini, C.; Scita, G.; et al. L-Proline induces a mesenchymal-like invasive program in embryonic stem cells by remodeling H3K9 and H3K36 methylation. Stem Cell Rep. 2013, 1, 307–321. [Google Scholar] [CrossRef]
- D’Aniello, C.; Cermola, F.; Patriarca, E.J.; Minchiotti, G. Vitamin C in Stem Cell Biology: Impact on Extracellular Matrix Homeostasis and Epigenetics. Stem Cells Int. 2017, 2017, 8936156. [Google Scholar] [CrossRef]
- Casalino, L.; Comes, S.; Lambazzi, G.; De Stefano, B.; Filosa, S.; De Falco, S.; De Cesare, D.; Minchiotti, G.; Patriarca, E.J. Control of embryonic stem cell metastability by L-proline catabolism. J. Mol. Cell Biol. 2011, 3, 108–122. [Google Scholar] [CrossRef]
- Washington, J.M.; Rathjen, J.; Felquer, F.; Lonic, A.; Bettess, M.D.; Hamra, N.; Semendric, L.; Tan, B.S.; Lake, J.A.; Keough, R.A.; et al. L-Proline induces differentiation of ES cells: A novel role for an amino acid in the regulation of pluripotent cells in culture. Am. J. Physiol. Cell Physiol. 2010, 298, C982–C992. [Google Scholar] [CrossRef]
- D’Aniello, C.; Fico, A.; Casalino, L.; Guardiola, O.; Di Napoli, G.; Cermola, F.; De Cesare, D.; Tate, R.; Cobellis, G.; Patriarca, E.J.; et al. A novel autoregulatory loop between the Gcn2-Atf4 pathway and (L)-Proline [corrected] metabolism controls stem cell identity. Cell Death Differ. 2015, 22, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, C.; Habibi, E.; Cermola, F.; Paris, D.; Russo, F.; Fiorenzano, A.; Di Napoli, G.; Melck, D.J.; Cobellis, G.; Angelini, C.; et al. Vitamin C and l-Proline Antagonistic Effects Capture Alternative States in the Pluripotency Continuum. Stem Cell Rep. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, C.; Cermola, F.; Palamidessi, A.; Wanderlingh, L.G.; Gagliardi, M.; Migliaccio, A.; Varrone, F.; Casalino, L.; Matarazzo, M.R.; De Cesare, D.; et al. Collagen prolyl hydroxylation-dependent metabolic perturbation governs epigenetic remodeling and mesenchymal transition in pluripotent and cancer cells. Cancer Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, Y.; Huber, W.; Tsumura, A.; Kang, M.; Xenopoulos, P.; Kurimoto, K.; Oles, A.K.; Arauzo-Bravo, M.J.; Saitou, M.; Hadjantonakis, A.K.; et al. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat. Cell Biol. 2014, 16, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Carey, B.W.; Finley, L.W.; Cross, J.R.; Allis, C.D.; Thompson, C.B. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2015, 518, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.Y.; Kwak, S.; Lee, S.; Kim, H.; Lee, S.E.; Kim, J.H.; Kim, Y.A.; Jeon, Y.K.; Chung, D.H.; Jin, X.; et al. Psat1-Dependent Fluctuations in alpha-Ketoglutarate Affect the Timing of ESC Differentiation. Cell Metab. 2016, 24, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Tischler, J.; Gruhn, W.H.; Reid, J.; Allgeyer, E.; Buettner, F.; Marr, C.; Theis, F.; Simons, B.D.; Wernisch, L.; Surani, M.A. Metabolic regulation of pluripotency and germ cell fate through alpha-ketoglutarate. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- TeSlaa, T.; Chaikovsky, A.C.; Lipchina, I.; Escobar, S.L.; Hochedlinger, K.; Huang, J.; Graeber, T.G.; Braas, D.; Teitell, M.A. alpha-Ketoglutarate Accelerates the Initial Differentiation of Primed Human Pluripotent Stem Cells. Cell Metab. 2016, 24, 485–493. [Google Scholar] [CrossRef]
- Zhang, Z.; He, C.; Zhang, L.; Zhu, T.; Lv, D.; Li, G.; Song, Y.; Wang, J.; Wu, H.; Ji, P.; et al. Alpha-ketoglutarate affects murine embryo development through metabolic and epigenetic modulations. Reproduction 2019, 158, 125–135. [Google Scholar] [CrossRef]
- Cimmino, L.; Neel, B.G.; Aifantis, I. Vitamin C in Stem Cell Reprogramming and Cancer. Trends Cell Biol. 2018, 28, 698–708. [Google Scholar] [CrossRef]
- Esteban, M.A.; Wang, T.; Qin, B.; Yang, J.; Qin, D.; Cai, J.; Li, W.; Weng, Z.; Chen, J.; Ni, S.; et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 2010, 6, 71–79. [Google Scholar] [CrossRef]
- Esteban, M.A.; Pei, D. Vitamin C improves the quality of somatic cell reprogramming. Nat. Genet. 2012, 44, 366–367. [Google Scholar] [CrossRef]
- Wang, T.; Chen, K.; Zeng, X.; Yang, J.; Wu, Y.; Shi, X.; Qin, B.; Zeng, L.; Esteban, M.A.; Pan, G.; et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 2011, 9, 575–587. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H.; Liu, J.; Qi, J.; Wei, B.; Yang, J.; Liang, H.; Chen, Y.; Chen, J.; Wu, Y.; et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013, 45, 34–42. [Google Scholar] [CrossRef]
- Stadtfeld, M.; Apostolou, E.; Ferrari, F.; Choi, J.; Walsh, R.M.; Chen, T.; Ooi, S.S.; Kim, S.Y.; Bestor, T.H.; Shioda, T.; et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012, 44, 398–405. [Google Scholar] [CrossRef]
- Xu, X.; Smorag, L.; Nakamura, T.; Kimura, T.; Dressel, R.; Fitzner, A.; Tan, X.; Linke, M.; Zechner, U.; Engel, W.; et al. Dppa3 expression is critical for generation of fully reprogrammed iPS cells and maintenance of Dlk1-Dio3 imprinting. Nat. Commun 2015, 6, 6008. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Han, Z.; Li, Q.; Wu, Y.; Shi, X.; Ai, Z.; Du, J.; Li, W.; Guo, Z.; Zhang, Y. Vitamin C induces a pluripotent state in mouse embryonic stem cells by modulating microRNA expression. Febs. J. 2015, 282, 685–699. [Google Scholar] [CrossRef]
- Doege, C.A.; Inoue, K.; Yamashita, T.; Rhee, D.B.; Travis, S.; Fujita, R.; Guarnieri, P.; Bhagat, G.; Vanti, W.B.; Shih, A.; et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 2012, 488, 652–655. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Guo, L.; Zhang, L.; Wu, H.; Yang, J.; Liu, H.; Wang, X.; Hu, X.; Gu, T.; Zhou, Z.; et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 2013, 45, 1504–1509. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, J.; Li, K.; Wu, T.; Huang, B.; Liu, W.; Kou, X.; Zhang, Y.; Huang, H.; Jiang, Y.; et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 2013, 12, 453–469. [Google Scholar] [CrossRef]
- Blaschke, K.; Ebata, K.T.; Karimi, M.M.; Zepeda-Martinez, J.A.; Goyal, P.; Mahapatra, S.; Tam, A.; Laird, D.J.; Hirst, M.; Rao, A.; et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 2013, 500, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yang, L.; Chen, L.; Wang, X.; Wu, H.; Ai, Z.; Du, J.; Liu, Y.; Shi, X.; Wu, Y.; et al. Vitamin C facilitates pluripotent stem cell maintenance by promoting pluripotency gene transcription. Biochimie 2013, 95, 2107–2113. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.L.; Brena, R.M.; Kolle, G.; Grimmond, S.M.; Berman, B.P.; Laird, P.W.; Pera, M.F.; Wolvetang, E.J. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells 2010, 28, 1848–1855. [Google Scholar] [CrossRef]
- Hore, T.A.; von Meyenn, F.; Ravichandran, M.; Bachman, M.; Ficz, G.; Oxley, D.; Santos, F.; Balasubramanian, S.; Jurkowski, T.P.; Reik, W. Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naive pluripotency by complementary mechanisms. Proc. Natl. Acad. Sci. USA 2016, 113, 12202–12207. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.; Teissandier, A.; Perez-Palacios, R.; Bourc’his, D. An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Dawlaty, M.M.; Breiling, A.; Le, T.; Barrasa, M.I.; Raddatz, G.; Gao, Q.; Powell, B.E.; Cheng, A.W.; Faull, K.F.; Lyko, F.; et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 2014, 29, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Langlois, T.; da Costa Reis Monte-Mor, B.; Lenglet, G.; Droin, N.; Marty, C.; Le Couedic, J.P.; Almire, C.; Auger, N.; Mercher, T.; Delhommeau, F.; et al. TET2 deficiency inhibits mesoderm and hematopoietic differentiation in human embryonic stem cells. Stem Cells 2014, 32, 2084–2097. [Google Scholar] [CrossRef] [PubMed]
- Norman, L.; Tarrant, P.; Chevassut, T. TET2 Inhibits Differentiation of Embryonic Stem Cells but Does Not Overcome Methylation-Induced Gene Silencing. Bone Marrow Res. 2014, 2014, 986571. [Google Scholar] [CrossRef] [PubMed]
- Dawlaty, M.M.; Breiling, A.; Le, T.; Raddatz, G.; Barrasa, M.I.; Cheng, A.W.; Gao, Q.; Powell, B.E.; Li, Z.; Xu, M.; et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 2013, 24, 310–323. [Google Scholar] [CrossRef]
- Koh, K.P.; Yabuuchi, A.; Rao, S.; Huang, Y.; Cunniff, K.; Nardone, J.; Laiho, A.; Tahiliani, M.; Sommer, C.A.; Mostoslavsky, G.; et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011, 8, 200–213. [Google Scholar] [CrossRef]
- Kang, J.; Lienhard, M.; Pastor, W.A.; Chawla, A.; Novotny, M.; Tsagaratou, A.; Lasken, R.S.; Thompson, E.C.; Surani, M.A.; Koralov, S.B.; et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E4236–E4245. [Google Scholar] [CrossRef] [Green Version]
- Ross, S.E.; Bogdanovic, O. TET enzymes, DNA demethylation and pluripotency. Biochem Soc Trans. 2019, 47, 875–885. [Google Scholar] [CrossRef]
- Takahashi, T.; Lord, B.; Schulze, P.C.; Fryer, R.M.; Sarang, S.S.; Gullans, S.R.; Lee, R.T. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation 2003, 107, 1912–1916. [Google Scholar] [CrossRef]
- Crescini, E.; Gualandi, L.; Uberti, D.; Prandelli, C.; Presta, M.; Dell’Era, P. Ascorbic acid rescues cardiomyocyte development in Fgfr1(-/-) murine embryonic stem cells. Biochim Biophys Acta 2013, 1833, 140–147. [Google Scholar] [CrossRef]
- Cao, N.; Liu, Z.; Chen, Z.; Wang, J.; Chen, T.; Zhao, X.; Ma, Y.; Qin, L.; Kang, J.; Wei, B.; et al. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012, 22, 219–236. [Google Scholar] [CrossRef]
- Buttery, L.D.; Bourne, S.; Xynos, J.D.; Wood, H.; Hughes, F.J.; Hughes, S.P.; Episkopou, V.; Polak, J.M. Differentiation of osteoblasts and in vitro bone formation from murine embryonic stem cells. Tissue Eng 2001, 7, 89–99. [Google Scholar] [CrossRef]
- Tsuneto, M.; Yamazaki, H.; Yoshino, M.; Yamada, T.; Hayashi, S. Ascorbic acid promotes osteoclastogenesis from embryonic stem cells. Biochem. Biophys. Res. Commun 2005, 335, 1239–1246. [Google Scholar] [CrossRef]
- Cuaranta-Monroy, I.; Simandi, Z.; Kolostyak, Z.; Doan-Xuan, Q.M.; Poliska, S.; Horvath, A.; Nagy, G.; Bacso, Z.; Nagy, L. Highly efficient differentiation of embryonic stem cells into adipocytes by ascorbic acid. Stem Cell Res. 2014, 13, 88–97. [Google Scholar] [CrossRef] [Green Version]
- Garcia, B.A.; Luka, Z.; Loukachevitch, L.V.; Bhanu, N.V.; Wagner, C. Folate deficiency affects histone methylation. Med. Hypotheses 2016, 88, 63–67. [Google Scholar] [CrossRef]
- Mentch, S.J.; Mehrmohamadi, M.; Huang, L.; Liu, X.; Gupta, D.; Mattocks, D.; Gomez Padilla, P.; Ables, G.; Bamman, M.M.; Thalacker-Mercer, A.E.; et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015, 22, 861–873. [Google Scholar] [CrossRef] [Green Version]
- Benayoun, B.A.; Pollina, E.A.; Ucar, D.; Mahmoudi, S.; Karra, K.; Wong, E.D.; Devarajan, K.; Daugherty, A.C.; Kundaje, A.B.; Mancini, E.; et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 2014, 158, 673–688. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M.; Ali, M.M. Methyl Donor Micronutrients that Modify DNA Methylation and Cancer Outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.; Martinez-Fernandez, A.; Faustino, R.S.; Yamada, S.; Perez-Terzic, C.; Nelson, T.J.; Terzic, A. Nuclear reprogramming with c-Myc potentiates glycolytic capacity of derived induced pluripotent stem cells. J. Cardiovasc. Transl. Res. 2013, 6, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Guo, W.T.; Tian, S.; He, X.; Wang, X.W.; Liu, X.; Gu, K.L.; Ma, X.; Huang, D.; Hu, L.; et al. miR-290/371-Mbd2-Myc circuit regulates glycolytic metabolism to promote pluripotency. EMBO J. 2015, 34, 609–623. [Google Scholar] [CrossRef]
- Mathieu, J.; Zhou, W.; Xing, Y.; Sperber, H.; Ferreccio, A.; Agoston, Z.; Kuppusamy, K.T.; Moon, R.T.; Ruohola-Baker, H. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell 2014, 14, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Fiorenzano, A.; Pascale, E.; D’Aniello, C.; Acampora, D.; Bassalert, C.; Russo, F.; Andolfi, G.; Biffoni, M.; Francescangeli, F.; Zeuner, A.; et al. Cripto is essential to capture mouse epiblast stem cell and human embryonic stem cell pluripotency. Nat. Commun. 2016, 7, 12589. [Google Scholar] [CrossRef] [PubMed]
- Carbognin, E.; Betto, R.M.; Soriano, M.E.; Smith, A.G.; Martello, G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 2016, 35, 618–634. [Google Scholar] [CrossRef] [PubMed]
- Sone, M.; Morone, N.; Nakamura, T.; Tanaka, A.; Okita, K.; Woltjen, K.; Nakagawa, M.; Heuser, J.E.; Yamada, Y.; Yamanaka, S.; et al. Hybrid Cellular Metabolism Coordinated by Zic3 and Esrrb Synergistically Enhances Induction of Naive Pluripotency. Cell Metab. 2017, 25, 1103–1117 e1106. [Google Scholar] [CrossRef]
- Kida, Y.S.; Kawamura, T.; Wei, Z.; Sogo, T.; Jacinto, S.; Shigeno, A.; Kushige, H.; Yoshihara, E.; Liddle, C.; Ecker, J.R.; et al. ERRs Mediate a Metabolic Switch Required for Somatic Cell Reprogramming to Pluripotency. Cell Stem Cell 2015, 16, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Ratanasirintrawoot, S.; Chandrasekaran, S.; Wu, Z.; Ficarro, S.B.; Yu, C.; Ross, C.A.; Cacchiarelli, D.; Xia, Q.; Seligson, M.; et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell 2016, 19, 66–80. [Google Scholar] [CrossRef] [Green Version]
- Marsboom, G.; Zhang, G.F.; Pohl-Avila, N.; Zhang, Y.; Yuan, Y.; Kang, H.; Hao, B.; Brunengraber, H.; Malik, A.B.; Rehman, J. Glutamine Metabolism Regulates the Pluripotency Transcription Factor OCT4. Cell Rep. 2016, 16, 323–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houghton, F.D.; Thompson, J.G.; Kennedy, C.J.; Leese, H.J. Oxygen consumption and energy metabolism of the early mouse embryo. Mol. Reprod. Dev. 1996, 44, 476–485. [Google Scholar] [CrossRef]
- Mathieu, J.; Ruohola-Baker, H. Metabolic remodeling during the loss and acquisition of pluripotency. Development 2017, 144, 541–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boroviak, T.; Loos, R.; Lombard, P.; Okahara, J.; Behr, R.; Sasaki, E.; Nichols, J.; Smith, A.; Bertone, P. Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Dev. Cell 2015, 35, 366–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaraj, R.; Sharpley, M.S.; Chi, F.; Braas, D.; Zhou, Y.; Kim, R.; Clark, A.T.; Banerjee, U. Nuclear Localization of Mitochondrial TCA Cycle Enzymes as a Critical Step in Mammalian Zygotic Genome Activation. Cell 2017, 168, 210–223 e211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, J.; Dahan, P.; Lu, V.; Zhang, C.; Li, H.; Teitell, M.A. Metabolism in Pluripotent Stem Cells and Early Mammalian Development. Cell Metab. 2018, 27, 332–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scognamiglio, R.; Cabezas-Wallscheid, N.; Thier, M.C.; Altamura, S.; Reyes, A.; Prendergast, A.M.; Baumgartner, D.; Carnevalli, L.S.; Atzberger, A.; Haas, S.; et al. Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell 2016, 164, 668–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulut-Karslioglu, A.; Biechele, S.; Jin, H.; Macrae, T.A.; Hejna, M.; Gertsenstein, M.; Song, J.S.; Ramalho-Santos, M. Inhibition of mTOR induces a paused pluripotent state. Nature 2016, 540, 119–123. [Google Scholar] [CrossRef] [Green Version]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. Methyltransferases modulate RNA stability in embryonic stem cells. Nat. Cell Biol. 2014, 16, 129–131. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Aniello, C.; Cermola, F.; Patriarca, E.J.; Minchiotti, G. Metabolic–Epigenetic Axis in Pluripotent State Transitions. Epigenomes 2019, 3, 13. https://0-doi-org.brum.beds.ac.uk/10.3390/epigenomes3030013
D’Aniello C, Cermola F, Patriarca EJ, Minchiotti G. Metabolic–Epigenetic Axis in Pluripotent State Transitions. Epigenomes. 2019; 3(3):13. https://0-doi-org.brum.beds.ac.uk/10.3390/epigenomes3030013
Chicago/Turabian StyleD’Aniello, Cristina, Federica Cermola, Eduardo J. Patriarca, and Gabriella Minchiotti. 2019. "Metabolic–Epigenetic Axis in Pluripotent State Transitions" Epigenomes 3, no. 3: 13. https://0-doi-org.brum.beds.ac.uk/10.3390/epigenomes3030013