Epigenetics of Skeletal Muscle-Associated Genes in the ASB, LRRC, TMEM, and OSBPL Gene Families
Abstract
:1. Introduction
2. Results
2.1. The SkM-Related Genes and Their Gene Families Chosen for Epigenetic Analysis
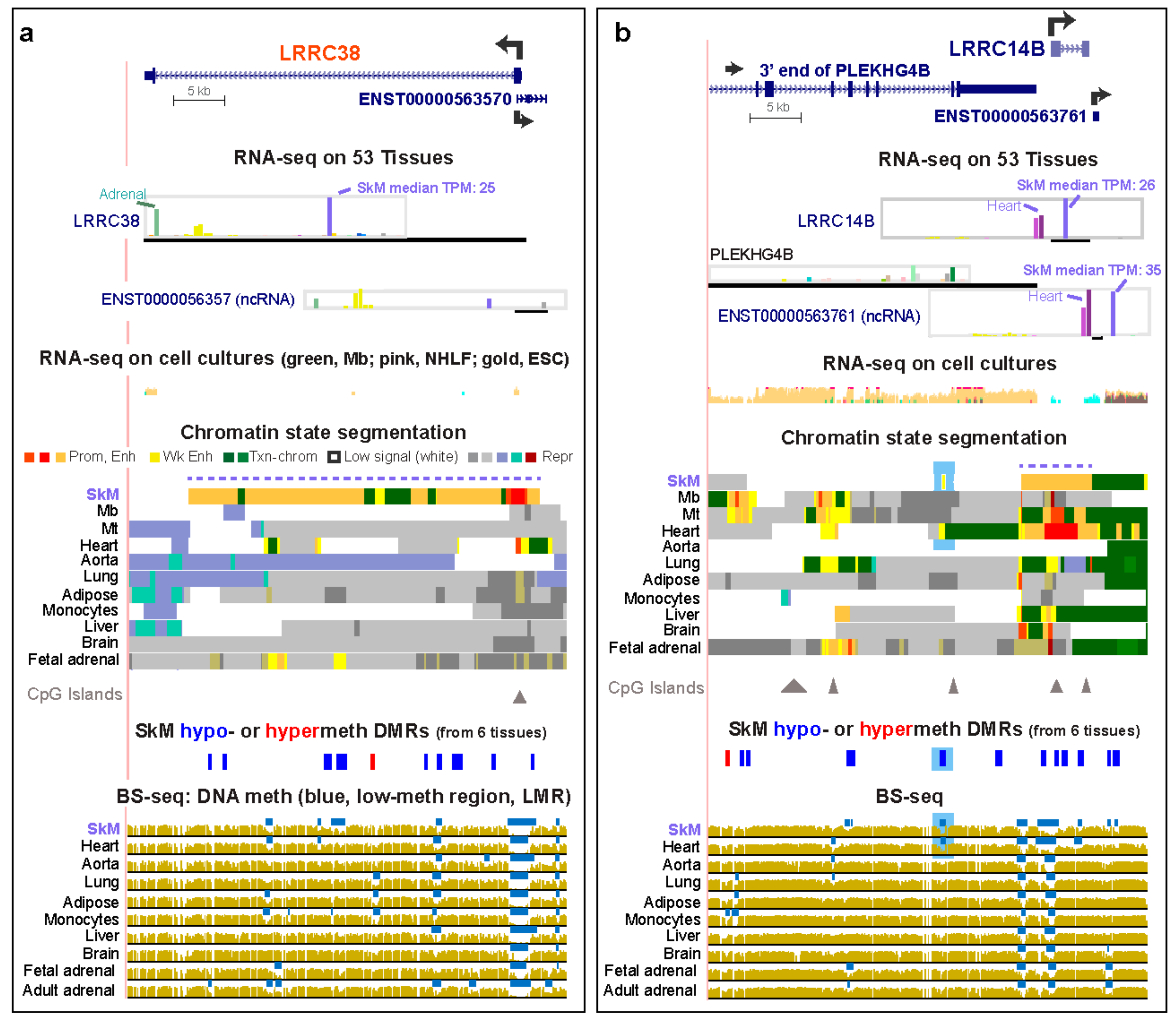
2.2. LRRC Genes that are Preferentially Expressed in SkM Display Muscle-Associated Enhancers or Super-Enhancers Containing Hypomethylated DNA Regions
2.3. Preferential Expression of OSBPL6 and OSBPL11 in SkM was Associated with SkM-Specific Enhancer Chromatin Mostly within the Gene Body or Upstream
2.4. Nine of the 18 ASB Genes Display SkM or SkM/Heart Preferential Expression that was Associated with Loss of DNA Methylation in or Adjacent to the Promoter
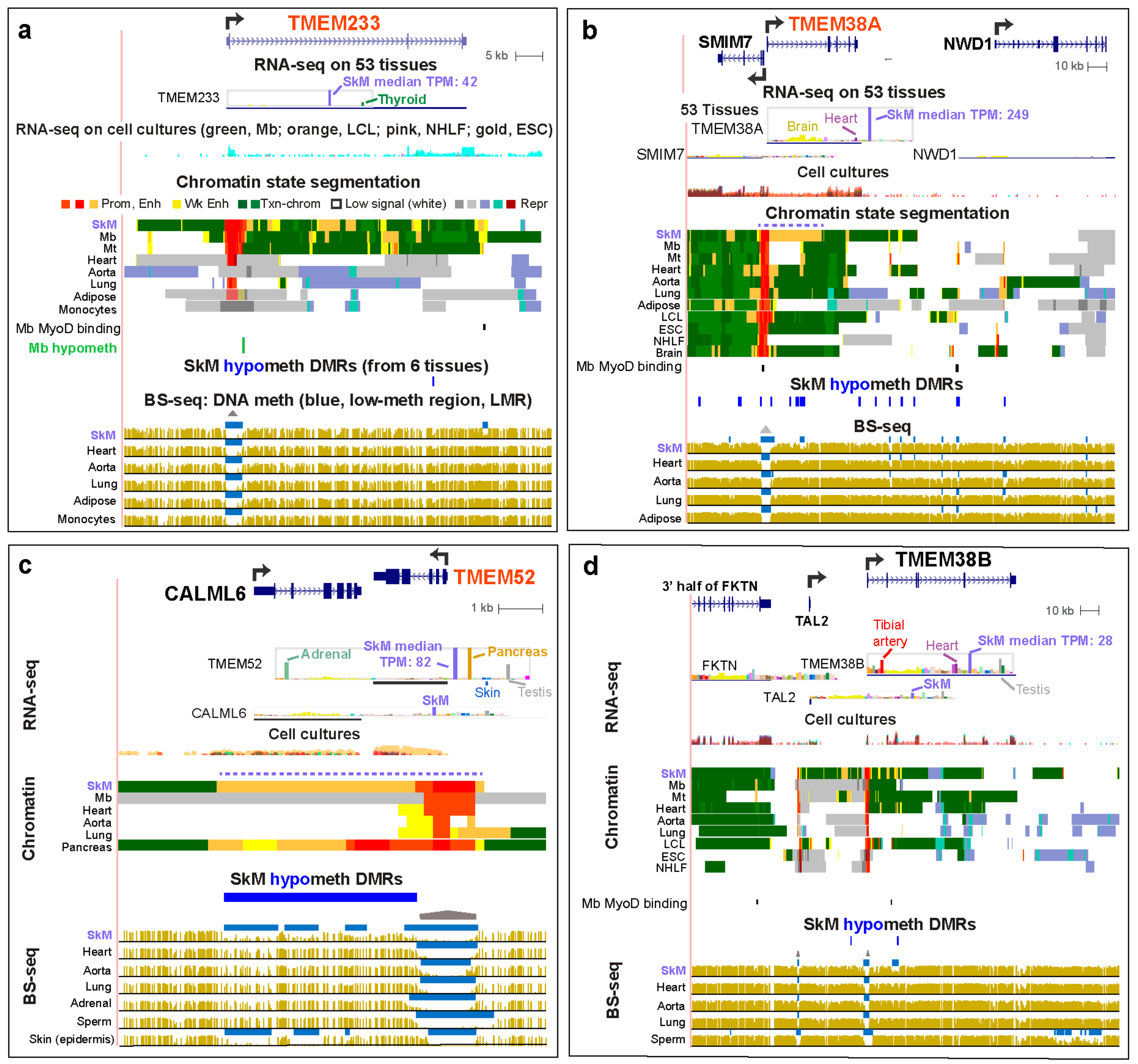
2.5. SkM-Associated Genes in the TMEM Family Exhibited SkM-Specific Intragenic, Intergenic, or Neighboring-Gene Enhancer Chromatin
2.6. Related Sequences of the Proteins Encoded by Some of the Studied SkM or SkM/Heart Genes Suggest Functional Similarities
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gonzalez-Freire, M.; Semba, R.D.; Ubaida-Mohien, C.; Fabbri, E.; Scalzo, P.; Hojlund, K.; Dufresne, C.; Lyashkov, A.; Ferrucci, L. The Human Skeletal Muscle Proteome Project: A reappraisal of the current literature. J. Cachexia Sarcopenia Muscle 2017, 8, 5–18. [Google Scholar] [CrossRef]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O’Brien, T.D.; Someren, K.A.V.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Methylome of human skeletal muscle after acute & chronic resistance exercise training, detraining & retraining. Sci. Data 2018, 5, 180213. [Google Scholar] [PubMed] [Green Version]
- Widmann, M.; Niess, A.M.; Munz, B. Physical exercise and epigenetic modifications in skeletal muscle. Sports Med. 2019, 49, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Lindskog, C.; Linne, J.; Fagerberg, L.; Hallstrom, B.M.; Sundberg, C.J.; Lindholm, M.; Huss, M.; Kampf, C.; Choi, H.; Liem, D.A.; et al. The human cardiac and skeletal muscle proteomes defined by transcriptomics and antibody-based profiling. BMC Genom. 2015, 16, 475. [Google Scholar] [CrossRef] [PubMed]
- Terry, E.E.; Zhang, X.; Hoffmann, C.; Hughes, L.D.; Lewis, S.A.; Li, J.; Wallace, M.J.; Riley, L.A.; Douglas, C.M.; Gutierrez-Monreal, M.A.; et al. Transcriptional profiling reveals extraordinary diversity among skeletal muscle tissues. Elife 2018, 7. [Google Scholar]
- The_GTEx_Consortium. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.W.; Mahmood, S.; Srinivasan, M.; Smiraglia, D.J.; Patel, M.S. Developmental programming in skeletal muscle in response to overnourishment in the immediate postnatal life in rats. J. Nutr. Biochem. 2013, 24, 1859–1869. [Google Scholar] [CrossRef] [Green Version]
- Tsumagari, K.; Baribault, C.; Terragni, J.; Chandra, S.; Renshaw, C.; Sun, Z.; Song, L.; Crawford, G.E.; Pradhan, S.; Lacey, M.; et al. DNA methylation and differentiation: HOX genes in muscle cells. Epigen. Chromatin 2013, 6, 25. [Google Scholar] [CrossRef] [Green Version]
- Segales, J.; Perdiguero, E.; Munoz-Canoves, P. Epigenetic control of adult skeletal muscle stem cell functions. FEBS J. 2015, 282, 1571–1588. [Google Scholar] [CrossRef] [Green Version]
- Tsumagari, K.; Baribault, C.; Terragni, J.; Varley, K.E.; Gertz, J.; Pradhan, S.; Baddoo, M.; Crain, C.M.; Song, L.; Crawford, G.E.; et al. Early de novo DNA methylation and prolonged demethylation in the muscle lineage. Epigenetics 2013, 8, 317–332. [Google Scholar] [CrossRef] [Green Version]
- Terragni, J.; Zhang, G.; Sun, Z.; Pradhan, S.; Song, L.; Crawford, G.E.; Lacey, M.; Ehrlich, M. Notch signaling genes: Myogenic DNA hypomethylation and 5-hydroxymethylcytosine. Epigenetics 2014, 9, 842–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begue, G.; Raue, U.; Jemiolo, B.; Trappe, S. DNA methylation assessment from human slow- and fast-twitch skeletal muscle fibers. J. Appl. Physiol. (1985) 2017, 122, 952–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wardle, F.C. Master control: Transcriptional regulation of mammalian Myod. J. Muscle Res. Cell Motil. 2019, 40, 211–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrio, E.; Diez-Villanueva, A.; Lois, S.; Mallona, I.; Cases, I.; Forn, M.; Peinado, M.A.; Suelves, M. Deconstruction of DNA methylation patterns during myogenesis reveals specific epigenetic events in the establishment of the skeletal muscle lineage. Stem Cells 2015, 33, 2025–2036. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Terragni, J.; Zhang, G.; Pradhan, S.; Haushka, S.; Johnston, D.; Baribault, C.; Lacey, M.; Ehrlich, M. Tissue-specific epigenetics in gene neighborhoods: Myogenic transcription factor genes. Hum. Mol. Genet. 2015, 24, 4660–4673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrlich, K.C.; Paterson, H.L.; Lacey, M.; Ehrlich, M. DNA hypomethylation in intragenic and intergenic enhancer chromatin of muscle-specific genes usually correlates with their expression. Yale J. Biol. Med. 2016, 89, 441–455. [Google Scholar] [PubMed]
- Asp, P.; Blum, R.; Vethantham, V.; Parisi, F.; Micsinai, M.; Cheng, J.; Bowman, C.; Kluger, Y.; Dynlacht, B.D. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. USA 2011, 108, E149–E158. [Google Scholar] [CrossRef] [Green Version]
- Moresi, V.; Marroncelli, N.; Coletti, D.; Adamo, S. Regulation of skeletal muscle development and homeostasis by gene imprinting, histone acetylation and microRNA. Biochim. Biophys. Acta 2015, 1849, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Blum, R. Activation of muscle enhancers by MyoD and epigenetic modifiers. J. Cell Biochem. 2014. [Google Scholar] [CrossRef]
- Marino, S.; Di Foggia, V. Invited Review: Polycomb group genes in the regeneration of the healthy and pathological skeletal muscle. Neuropathol. Appl. Neurobiol. 2016, 42, 407–422. [Google Scholar] [CrossRef]
- Wang, Y.; Hao, Y.; Alway, S.E. Suppression of GSK-3beta activation by M-cadherin protects myoblasts against mitochondria-associated apoptosis during myogenic differentiation. J. Cell Sci. 2011, 124, 3835–3847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhanu, N.V.; Sidoli, S.; Yuan, Z.F.; Molden, R.C.; Garcia, B.A. Regulation of proline-directed kinases and the trans-histone code H3K9me3/H4K20me3 during human myogenesis. J. Biol. Chem. 2019, 294, 8296–8308. [Google Scholar] [CrossRef] [PubMed]
- Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; Ziller, M.J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; So, K.K.; Li, Y.; Li, Y.; Yuan, J.; Ding, Y.; Chen, F.; Huang, Y.; Liu, J.; Lee, W.; et al. Elevated H3K27ac in aged skeletal muscle leads to increase in extracellular matrix and fibrogenic conversion of muscle satellite cells. Aging Cell 2019, 18, e12996. [Google Scholar] [CrossRef] [Green Version]
- Breuls, N.; Giacomazzi, G.; Sampaolesi, M. (Epi)genetic modifications in myogenic stem cells: From novel insights to therapeutic perspectives. Cells 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Zhang, X. dbSUPER: A database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016, 44, D164–D171. [Google Scholar] [CrossRef] [Green Version]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-Andre, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-enhancers in the control of cell identity and disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Decato, B.; Hong, E.E.; Zhou, M.; Fang, F.; Qu, J.; Garvin, T.; Kessler, M.; Zhou, J.; Smith, A.D. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS ONE 2013, 8, e81148. [Google Scholar] [CrossRef] [Green Version]
- Will, R.D.; Eden, M.; Just, S.; Hansen, A.; Eder, A.; Frank, D.; Kuhn, C.; Seeger, T.S.; Oehl, U.; Wiemann, S.; et al. Myomasp/LRRC39, a heart- and muscle-specific protein, is a novel component of the sarcomeric M-band and is involved in stretch sensing. Circ. Res. 2010, 107, 1253–1264. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Perez, V.; Lingle, C.J. Regulation of BK channels by beta and bamma subunits. Annu. Rev. Physiol. 2019, 81, 113–137. [Google Scholar] [CrossRef]
- McDermott-Roe, C.; Leleu, M.; Rowe, G.C.; Palygin, O.; Bukowy, J.D.; Kuo, J.; Rech, M.; Hermans-Beijnsberger, S.; Schaefer, S.; Adami, E.; et al. Transcriptome-wide co-expression analysis identifies LRRC2 as a novel mediator of mitochondrial and cardiac function. PLoS ONE 2017, 12, e0170458. [Google Scholar] [CrossRef] [PubMed]
- Larrouy, D.; Barbe, P.; Valle, C.; Dejean, S.; Pelloux, V.; Thalamas, C.; Bastard, J.P.; Le Bouil, A.; Diquet, B.; Clement, K.; et al. Gene expression profiling of human skeletal muscle in response to stabilized weight loss. Am. J. Clin. Nutr. 2008, 88, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Jin, J.; Xu, Z.; Zuo, B. Functions and regulatory mechanisms of lncRNAs in skeletal myogenesis, muscle disease and meat production. Cells 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Haeussler, M.; Zweig, A.S.; Tyner, C.; Speir, M.L.; Rosenbloom, K.R.; Raney, B.J.; Lee, C.M.; Lee, B.T.; Hinrichs, A.S.; Gonzalez, J.N.; et al. The UCSC Genome Browser database: 2019 update. Nucleic. Acids Res. 2019, 47, D853–D858. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Yao, Z.; Sarkar, D.; Lawrence, M.; Sanchez, G.J.; Parker, M.H.; MacQuarrie, K.L.; Davison, J.; Morgan, M.T.; Ruzzo, W.L.; et al. Genome-wide MyoD binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev. Cell 2010, 18, 662–674. [Google Scholar] [CrossRef] [Green Version]
- Kentala, H.; Weber-Boyvat, M.; Olkkonen, V.M. OSBP-related protein family: Mediators of lipid transport and signaling at membrane contact sites. Int. Rev. Cell Mol. Biol. 2016, 321, 299–340. [Google Scholar]
- Liu, P.; Verhaar, A.P.; Peppelenbosch, M.P. Signaling size: Ankyrin and SOCS Box-Containing ASB E3 ligases in action. Trends Biochem. Sci. 2019, 44, 64–74. [Google Scholar] [CrossRef]
- Anasa, V.V.; Ravanan, P.; Talwar, P. Multifaceted roles of ASB proteins and its pathological significance. Front. Biol. 2018, 13, 376–388. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, H.; Liu, J.; Mao, J. Long noncoding RNA ASB16-AS1 promotes proliferation, migration, and invasion in glioma cells. Biomed. Res. Int. 2019, 2019, 5437531. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, F.; Eberhardt, D.; Witschas, K.; El-Ajouz, S.; Iida, T.; Nishi, M.; Takeshima, H.; Sitsapesan, R.; Venturi, E. Enhanced activity of multiple TRIC-B channels: An endoplasmic reticulum/sarcoplasmic reticulum mechanism to boost counterion currents. J. Physiol. 2019, 597, 2691–2705. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.I.; de Las Heras, J.I.; Czapiewski, R.; Le Thanh, P.; Booth, D.G.; Kelly, D.A.; Webb, S.; Kerr, A.R.W.; Schirmer, E.C. Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Mol. Cell 2016, 62, 834–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, A.; Takeshima, H. TRIC-B mutations causing osteogenesis imperfecta. Biol. Pharm. Bull. 2016, 39, 1743–1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Guo, J.H.; Saiyin, H.; Chen, L.; Zhou, G.J.; Huang, C.Q.; Yu, L. Cloning and characterization of human CAGLP gene encoding a novel EF-hand protein. DNA Seq. 2004, 15, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Vihola, A.; Bachinski, L.L.; Sirito, M.; Olufemi, S.E.; Hajibashi, S.; Baggerly, K.A.; Raheem, O.; Haapasalo, H.; Suominen, T.; Holmlund-Hampf, J.; et al. Differences in aberrant expression and splicing of sarcomeric proteins in the myotonic dystrophies DM1 and DM2. Acta Neuropathol. 2010, 119, 465–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, T.; Komori, R.; Ishida, K.; Kino, K.; Tanuma, S.; Miyazawa, H. Tal2 expression is induced by all-trans retinoic acid in P19 cells prior to acquisition of neural fate. Sci. Rep. 2014, 4, 4935. [Google Scholar] [CrossRef]
- Courtial, N.; Mucke, C.; Herkt, S.; Kolodziej, S.; Hussong, H.; Lausen, J. The T-cell oncogene Tal2 Is a Target of PU.1 and upregulated during osteoclastogenesis. PLoS ONE 2013, 8, e76637. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Barger, C.J.; Eng, K.H.; Klinkebiel, D.; Link, P.A.; Omilian, A.; Bshara, W.; Odunsi, K.; Karpf, A.R. PRAME expression and promoter hypomethylation in epithelial ovarian cancer. Oncotarget 2016, 7, 45352–45369. [Google Scholar] [CrossRef] [Green Version]
- Woon, M.T.; Long, P.A.; Reilly, L.; Evans, J.M.; Keefe, A.M.; Lea, M.R.; Beglinger, C.J.; Balijepalli, R.C.; Lee, Y.; Olson, T.M.; et al. Pediatric dilated cardiomyopathy-associated LRRC10 (Leucine-Rich Repeat-Containing 10) variant reveals LRRC10 as an auxiliary subunit of xardiac L-type Ca(2+) channels. J. Am. Heart Assoc. 2018, 7. [Google Scholar]
- Pietrangelo, A.; Ridgway, N.D. Bridging the molecular and biological functions of the oxysterol-binding protein family. Cell Mol. Life Sci. 2018, 75, 3079–3098. [Google Scholar] [CrossRef]
- Lehto, M.; Laitinen, S.; Chinetti, G.; Johansson, M.; Ehnholm, C.; Staels, B.; Ikonen, E.; Olkkonen, V.M. The OSBP-related protein family in humans. J. Lipid Res. 2001, 42, 1203–1213. [Google Scholar] [PubMed]
- Benoit, G.; Warma, A.; Lussier, J.G.; Ndiaye, K. Gonadotropin regulation of ankyrin-repeat and SOCS-box protein 9 (ASB9) in ovarian follicles and identification of binding partners. PLoS ONE 2019, 14, e0212571. [Google Scholar] [CrossRef] [PubMed]
- Tee, J.M.; Peppelenbosch, M.P. Anchoring skeletal muscle development and disease: The role of ankyrin repeat domain containing proteins in muscle physiology. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 318–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazawa, M.; Ferrante, C.; Feng, J.; Mio, K.; Ogura, T.; Zhang, M.; Lin, P.H.; Pan, Z.; Komazaki, S.; Kato, K.; et al. TRIC channels are essential for Ca2+ handling in intracellular stores. Nature 2007, 448, 78–82. [Google Scholar] [CrossRef]
- Pitt, S.J.; Park, K.H.; Nishi, M.; Urashima, T.; Aoki, S.; Yamazaki, D.; Ma, J.; Takeshima, H.; Sitsapesan, R. Charade of the SR K+-channel: Two ion-channels, TRIC-A and TRIC-B, masquerade as a single K+-channel. Biophys. J. 2010, 99, 417–426. [Google Scholar] [CrossRef] [Green Version]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef]
- Barrientos, G.; Sanchez-Aguilera, P.; Jaimovich, E.; Hidalgo, C.; Llanos, P. Membrane cholesterol in skeletal muscle: A novel player in excitation-contraction coupling and insulin resistance. J. Diabetes Res. 2017, 2017, 3941898. [Google Scholar] [CrossRef]
- Kiss, H.; Yang, Y.; Kiss, C.; Andersson, K.; Klein, G.; Imreh, S.; Dumanski, J.P. The transcriptional map of the common eliminated region 1 (C3CER1) in 3p21.3. Eur. J. Hum. Genet. 2002, 10, 52–61. [Google Scholar] [CrossRef]
- Myers, R.M.; Stamatoyannopoulos, J.; Snyder, M.; Dunham, I.; Hardison, R.C.; Bernstein, B.E.; Gingeras, T.R.; Kent, W.J.; Birney, E.; Wold, B.; et al. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011, 9, e1001046. [Google Scholar]
- Okumura, F.; Joo-Okumura, A.; Nakatsukasa, K.; Kamura, T. The role of cullin 5-containing ubiquitin ligases. Cell Div. 2016, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Ishibashi, J.; Holterman, C.; Rudnicki, M.A. Muscle satellite cell-specific genes identified by genetic profiling of MyoD-deficient myogenic cell. Dev. Biol. 2004, 275, 287–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, J.H.; Conley, L.N.; Hedegaard, J.; Nielsen, M.; Young, J.F.; Oksbjerg, N.; Hornshoj, H.; Bendixen, C.; Thomsen, B. Gene expression profiling of porcine skeletal muscle in the early recovery phase following acute physical activity. Exp. Physiol. 2012, 97, 833–848. [Google Scholar] [CrossRef] [PubMed]
- Cappella, M.; Perfetti, A.; Cardinali, B.; Garcia-Manteiga, J.M.; Carrara, M.; Provenzano, C.; Fuschi, P.; Cardani, R.; Renna, L.V.; Meola, G.; et al. High-throughput analysis of the RNA-induced silencing complex in myotonic dystrophy type 1 patients identifies the dysregulation of miR-29c and its target ASB2. Cell Death Dis. 2018, 9, 729. [Google Scholar] [CrossRef] [PubMed]
- Kohroki, J.; Fujita, S.; Itoh, N.; Yamada, Y.; Imai, H.; Yumoto, N.; Nakanishi, T.; Tanaka, K. ATRA-regulated Asb-2 gene induced in differentiation of HL-60 leukemia cells. FEBS Lett. 2001, 505, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Bello, N.F.; Lamsoul, I.; Heuze, M.L.; Metais, A.; Moreaux, G.; Calderwood, D.A.; Duprez, D.; Moog-Lutz, C.; Lutz, P.G. The E3 ubiquitin ligase specificity subunit ASB2beta is a novel regulator of muscle differentiation that targets filamin B to proteasomal degradation. Cell Death Differ. 2009, 16, 921–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferguson, J.E., 3rd; Wu, Y.; Smith, K.; Charles, P.; Powers, K.; Wang, H.; Patterson, C. ASB4 is a hydroxylation substrate of FIH and promotes vascular differentiation via an oxygen-dependent mechanism. Mol. Cell Biol. 2007, 27, 6407–6419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Townley-Tilson, W.H.; Wu, Y.; Ferguson, J.E., 3rd; Patterson, C. The ubiquitin ligase ASB4 promotes trophoblast differentiation through the degradation of ID2. PLoS ONE 2014, 9, e89451. [Google Scholar] [CrossRef] [Green Version]
- McDaneld, T.G.; Spurlock, D.M. Ankyrin repeat and suppressor of cytokine signaling (SOCS) box-containing protein (ASB) 15 alters differentiation of mouse C2C12 myoblasts and phosphorylation of mitogen-activated protein kinase and Akt. J. Anim. Sci. 2008, 86, 2897–2902. [Google Scholar] [CrossRef]
- Boengler, K.; Pipp, F.; Fernandez, B.; Richter, A.; Schaper, W.; Deindl, E. The ankyrin repeat containing SOCS box protein 5: A novel protein associated with arteriogenesis. Biochem. Biophys. Res. Commun. 2003, 302, 17–22. [Google Scholar] [CrossRef]
- Thottakara, T.; Friedrich, F.W.; Reischmann, S.; Braumann, S.; Schlossarek, S.; Kramer, E.; Juhr, D.; Schluter, H.; van der Velden, J.; Munch, J.; et al. The E3 ubiquitin ligase Asb2beta is downregulated in a mouse model of hypertrophic cardiomyopathy and targets desmin for proteasomal degradation. J. Mol. Cell Cardiol. 2015, 87, 214–224. [Google Scholar] [CrossRef]
- Davey, J.R.; Watt, K.I.; Parker, B.L.; Chaudhuri, R.; Ryall, J.G.; Cunningham, L.; Qian, H.; Sartorelli, V.; Sandri, M.; Chamberlain, J.; et al. Integrated expression analysis of muscle hypertrophy identifies Asb2 as a negative regulator of muscle mass. JCI Insight 2016, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Yamazaki, D.; Park, K.H.; Komazaki, S.; Tjondrokoesoemo, A.; Nishi, M.; Lin, P.; Hirata, Y.; Brotto, M.; Takeshima, H.; et al. Ca2+ overload and sarcoplasmic reticulum instability in tric-a null skeletal muscle. J. Biol. Chem. 2010, 285, 37370–37376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Valle, I.; Buonocore, F.; Duncan, A.J.; Lin, L.; Barenco, M.; Parnaik, R.; Shah, S.; Hubank, M.; Gerrelli, D.; Achermann, J.C. A genomic atlas of human adrenal and gonad development. Wellcome Open Res. 2017, 2, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Aldrich, R.W. BK potassium channel modulation by leucine-rich repeat-containing proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 7917–7922. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Shimizu, N.; Yoshikawa, N. Role of skeletal muscle glucocorticoid receptor in systemic energy homeostasis. Exp. Cell Res. 2017, 360, 24–26. [Google Scholar] [CrossRef]
- Braun, T.P.; Marks, D.L. The regulation of muscle mass by endogenous glucocorticoids. Front. Physiol. 2015, 6, 12. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Lacey, M.R.; Baribault, C.; Ehrlich, M. Modeling, simulation and analysis of methylation profiles from reduced representation bisulfite sequencing experiments. Stat. Appl. Genet. Mol. Biol. 2013, 12, 723–742. [Google Scholar] [CrossRef]
- Lacey, M.; Baribault, C.; Ehrlich, K.C.; Ehrlich, M. Data showing atherosclerosis-associated differentially methylated regions are often at enhancers. Data Brief 2019, 23, 103812. [Google Scholar] [CrossRef]



| Epigenetic Patterns Associated with SkM | ASB Family (9 Genes) | LRRC Family (6 Genes) | OSBPL Family (2 Genes) | TMEM Family (4 Genes) |
|---|---|---|---|---|
| Prom chrom assoc with expression in SkM | ASB2, 4, 5, 10, 11, 15 | LRRC2, 14Bb, 20, 30, 38,39 | none | TMEM52, 233 |
| SkM-assoc prom DNA hypometh | ASB4, 5, 10, 11, 12, 15 | LRRC30, 39 | none | none |
| constit LMR at TSS | ASB2, 8, 16 | LRRC2, 14B, 20, 38 | OSBPL11 | none |
| CpG island at prom | ASB8 | LRRC14B, 20, 38 | OSBPL6, 11 | TMEM38A, 38B, 52 |
| Super-enhancer | ASB2,5, 8, 11 | LRRC14B, 20, 38 | none | TMEM52 |
| SkM enh chrom in adjacent gene | ASB2, 8, 10, 16 | LRRC14B, 39 | none | TMEM52; 38A |
| Intragenic enhancer chrom | ASB2, 4, 5, 8, 10, 11, 12, 15, 16 | LRRC2, 14B, 20, 38, 39 | OSBPL6, 11 | TMEM38A, 38B, 52, 233 |
| Intergenic proximal enh chrom | ASB2, 5, 8, 10, 12, 15, 16 | LRRC14B, 30, 38 | OSBPL11 | TMEM52 |
| Intergenic distal enh chrom | ASB2, 4, 10, 15, 16 | LRRC14B, 30 | OSBPL11 | TMEM38A, 38B, 233 |
| DNA hypometh in SkM-assoc enh chrom | ASB2, 5, 10, 11, 12, 15, 16 | LRRC2, 14B, 20, 30, 38, 39 | OSBPL6, 11 | TMEM38A, 38B, 52, 233 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehrlich, K.C.; Lacey, M.; Ehrlich, M. Epigenetics of Skeletal Muscle-Associated Genes in the ASB, LRRC, TMEM, and OSBPL Gene Families. Epigenomes 2020, 4, 1. https://0-doi-org.brum.beds.ac.uk/10.3390/epigenomes4010001
Ehrlich KC, Lacey M, Ehrlich M. Epigenetics of Skeletal Muscle-Associated Genes in the ASB, LRRC, TMEM, and OSBPL Gene Families. Epigenomes. 2020; 4(1):1. https://0-doi-org.brum.beds.ac.uk/10.3390/epigenomes4010001
Chicago/Turabian StyleEhrlich, Kenneth C., Michelle Lacey, and Melanie Ehrlich. 2020. "Epigenetics of Skeletal Muscle-Associated Genes in the ASB, LRRC, TMEM, and OSBPL Gene Families" Epigenomes 4, no. 1: 1. https://0-doi-org.brum.beds.ac.uk/10.3390/epigenomes4010001





