Epigenetic Regulators of White Adipocyte Browning
Abstract
:1. Introduction
2. Histone Acetyltransferases (HATs)
3. Histone Deacetylases (HDACs)
4. Histone Methyltransferases (HMT)
5. Histone Demethylases
6. DNA Methyltransferases and Demethylases
7. Transcriptional Factors of White Adipocytes and Browning
8. miRNA
9. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose Tissue Remodeling: Its Role in Energy Metabolism and Metabolic Disorders. Front. Endocrinol. 2016, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.B. Dynamic cross talk between metabolic organs in obesity and metabolic diseases. Exp. Mol. Med. 2016, 48, e214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegelman, B.M.; Flier, J.S. Obesity and the regulation of energy balance. Cell 2001, 104, 531–543. [Google Scholar] [CrossRef] [Green Version]
- Gesta, S.; Tseng, Y.H.; Kahn, C.R. Developmental origin of fat: Tracking obesity to its source. Cell 2007, 131, 242–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, D.; Picard, F. Brown fat biology and thermogenesis. Front. Biosci. (Landmark Ed.) 2011, 16, 1233–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Bostrom, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Berry, D.C.; Graff, J.M. Distinct cellular and molecular mechanisms for beta3 adrenergic receptor-induced beige adipocyte formation. eLife 2017, 6, e30329. [Google Scholar] [CrossRef]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scime, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Gurmaches, J.; Hung, C.M.; Sparks, C.A.; Tang, Y.; Li, H.; Guertin, D.A. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 2012, 16, 348–362. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Jun, H.; McDermott, J.R. Formation and activation of thermogenic fat. Trends Genet. 2015, 31, 232–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roh, H.C.; Tsai, L.T.Y.; Shao, M.; Tenen, D.; Shen, Y.; Kumari, M.; Lyubetskaya, A.; Jacobs, C.; Dawes, B.; Gupta, R.K.; et al. Warming Induces Significant Reprogramming of Beige, but Not Brown, Adipocyte Cellular Identity. Cell Metab. 2018, 27, 1121–1137. [Google Scholar] [CrossRef]
- Sears, M.R. Adverse effects of beta-agonists. J. Allergy Clin. Immunol 2002, 110, S322–S328. [Google Scholar] [CrossRef] [PubMed]
- Simona Negreș, C.C.; Arsene, A.L.; Margină, D.; Moroșan, E.; Zbârcea, C.E. New Potential Beta-3 Adrenergic Agonists with Beta- Phenylethylamine Structure, Synthesized for the Treatment of Dyslipidemia and Obesity. In Adiposity—Epidemiology and Treatment Modalities; IntechOpen: London, UK, 2017. [Google Scholar]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120. [Google Scholar] [CrossRef] [PubMed]
- Ong, B.X.; Brunmeir, R.; Zhang, Q.; Peng, X.; Idris, M.; Liu, C.; Xu, F. Regulation of Thermogenic Adipocyte Differentiation and Adaptive Thermogenesis through Histone Acetylation. Front. Endocrinol 2020, 11, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikkelsen, T.S.; Xu, Z.; Zhang, X.; Wang, L.; Gimble, J.M.; Lander, E.S.; Rosen, E.D. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010, 143, 156–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siersbaek, R.; Nielsen, R.; John, S.; Sung, M.H.; Baek, S.; Loft, A.; Hager, G.L.; Mandrup, S. Extensive chromatin remodelling and establishment of transcription factor ’hotspots’ during early adipogenesis. EMBO J. 2011, 30, 1459–1472. [Google Scholar] [CrossRef] [Green Version]
- Steger, D.J.; Grant, G.R.; Schupp, M.; Tomaru, T.; Lefterova, M.I.; Schug, J.; Manduchi, E.; Stoeckert, C.J., Jr.; Lazar, M.A. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010, 24, 1035–1044. [Google Scholar] [CrossRef] [Green Version]
- Jin, Q.; Yu, L.R.; Wang, L.; Zhang, Z.; Kasper, L.H.; Lee, J.E.; Wang, C.; Brindle, P.K.; Dent, S.Y.; Ge, K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011, 30, 249–262. [Google Scholar] [CrossRef]
- Takahashi, N.; Kawada, T.; Yamamoto, T.; Goto, T.; Taimatsu, A.; Aoki, N.; Kawasaki, H.; Taira, K.; Yokoyama, K.K.; Kamei, Y.; et al. Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 2002, 277, 16906–16912. [Google Scholar] [CrossRef] [Green Version]
- Na, H.H.; Kim, K.C. Homeostatic balance of histone acetylation and deconstruction of repressive chromatin marker H3K9me3 during adipocyte differentiation of 3T3-L1 cells. Genes Genom. 2018, 40, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Oike, Y.; Kamon, J.; Waki, H.; Komeda, K.; Tsuchida, A.; Date, Y.; Li, M.X.; Miki, H.; Akanuma, Y.; et al. Increased insulin sensitivity despite lipodystrophy in Crebbp heterozygous mice. Nat. Genet. 2002, 30, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Wang, C.; Kuang, X.; Feng, X.; Sartorelli, V.; Ying, H.; Ge, K.; Dent, S.Y. Gcn5 and PCAF regulate PPARgamma and Prdm16 expression to facilitate brown adipogenesis. Mol. Cell. Biol. 2014, 34, 3746–3753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawabe, Y.; Mori, J.; Morimoto, H.; Yamaguchi, M.; Miyagaki, S.; Ota, T.; Tsuma, Y.; Fukuhara, S.; Nakajima, H.; Oudit, G.Y.; et al. ACE2 exerts anti-obesity effect via stimulating brown adipose tissue and induction of browning in white adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E1140–E1149. [Google Scholar] [CrossRef]
- van Beekum, O.; Brenkman, A.B.; Grontved, L.; Hamers, N.; van den Broek, N.J.; Berger, R.; Mandrup, S.; Kalkhoven, E. The adipogenic acetyltransferase Tip60 targets activation function 1 of peroxisome proliferator-activated receptor gamma. Endocrinology 2008, 149, 1840–1849. [Google Scholar] [CrossRef]
- Picard, F.; Gehin, M.; Annicotte, J.; Rocchi, S.; Champy, M.F.; O’Malley, B.W.; Chambon, P.; Auwerx, J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 2002, 111, 931–941. [Google Scholar] [CrossRef] [Green Version]
- Tharkar-Promod, S.; Johnson, D.P.; Bennett, S.E.; Dennis, E.M.; Banowsky, B.G.; Jones, S.S.; Shearstone, J.R.; Quayle, S.N.; Min, C.; Jarpe, M.; et al. HDAC1,2 inhibition and doxorubicin impair Mre11-dependent DNA repair and DISC to override BCR-ABL1-driven DSB repair in Philadelphia chromosome-positive B-cell precursor acute lymphoblastic leukemia. Leukemia 2018, 32, 49–60. [Google Scholar] [CrossRef]
- Li, F.; Wu, R.; Cui, X.; Zha, L.; Yu, L.; Shi, H.; Xue, B. Histone Deacetylase 1 (HDAC1) Negatively Regulates Thermogenic Program in Brown Adipocytes via Coordinated Regulation of Histone H3 Lysine 27 (H3K27) Deacetylation and Methylation. J. Biol. Chem. 2016, 291, 4523–4536. [Google Scholar] [CrossRef] [Green Version]
- Di Giorgio, E.; Dalla, E.; Franforte, E.; Paluvai, H.; Minisini, M.; Trevisanut, M.; Picco, R.; Brancolini, C. Different class IIa HDACs repressive complexes regulate specific epigenetic responses related to cell survival in leiomyosarcoma cells. Nucleic Acids Res. 2020, 48, 646–664. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, A.; Longo, R.; Fiorino, E.; Silva, R.; Mitro, N.; Cermenati, G.; Gilardi, F.; Desvergne, B.; Andolfo, A.; Magagnotti, C.; et al. HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat. Commun. 2017, 8, 93. [Google Scholar] [CrossRef] [Green Version]
- Bagchi, R.A.; Ferguson, B.S.; Stratton, M.S.; Hu, T.; Cavasin, M.A.; Sun, L.; Lin, Y.H.; Liu, D.; Londono, P.; Song, K.; et al. HDAC11 suppresses the thermogenic program of adipose tissue via BRD2. JCI Insight 2018, 3, e120159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haberland, M.; Carrer, M.; Mokalled, M.H.; Montgomery, R.L.; Olson, E.N. Redundant control of adipogenesis by histone deacetylases 1 and 2. J. Biol. Chem. 2010, 285, 14663–14670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galmozzi, A.; Mitro, N.; Ferrari, A.; Gers, E.; Gilardi, F.; Godio, C.; Cermenati, G.; Gualerzi, A.; Donetti, E.; Rotili, D.; et al. Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes 2013, 62, 732–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, T.K.; Idelman, G.; Blanco, V.; Blomkalns, A.L.; Piegore, M.G., Jr.; Weintraub, D.S.; Kumar, S.; Rajsheker, S.; Manka, D.; Rudich, S.M.; et al. Histone deacetylase 9 is a negative regulator of adipogenic differentiation. J. Biol. Chem. 2011, 286, 27836–27847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, T.K.; Basford, J.E.; Knoll, E.; Tong, W.S.; Blanco, V.; Blomkalns, A.L.; Rudich, S.; Lentsch, A.B.; Hui, D.Y.; Weintraub, N.L. HDAC9 knockout mice are protected from adipose tissue dysfunction and systemic metabolic disease during high-fat feeding. Diabetes 2014, 63, 176–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rifai, K.; Judes, G.; Idrissou, M.; Daures, M.; Bignon, Y.J.; Penault-Llorca, F.; Bernard-Gallon, D. SIRT1-dependent epigenetic regulation of H3 and H4 histone acetylation in human breast cancer. Oncotarget 2018, 9, 30661–30678. [Google Scholar] [CrossRef]
- Bosch-Presegue, L.; Vaquero, A. Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J. 2015, 282, 1745–1767. [Google Scholar] [CrossRef]
- Vaquero, A.; Scher, M.B.; Lee, D.H.; Sutton, A.; Cheng, H.L.; Alt, F.W.; Serrano, L.; Sternglanz, R.; Reinberg, D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006, 20, 1256–1261. [Google Scholar] [CrossRef] [Green Version]
- Lai, Q.; Du, W.; Wu, J.; Wang, X.; Li, X.; Qu, X.; Wu, X.; Dong, F.; Yao, R.; Fan, H. H3K9ac and HDAC2 Activity Are Involved in the Expression of Monocarboxylate Transporter 1 in Oligodendrocyte. Front. Mol. Neurosci. 2017, 10, 376. [Google Scholar] [CrossRef]
- Zhou, Y.; Song, T.; Peng, J.; Zhou, Z.; Wei, H.; Zhou, R.; Jiang, S.; Peng, J. SIRT1 suppresses adipogenesis by activating Wnt/beta-catenin signaling in vivo and in vitro. Oncotarget 2016, 7, 77707–77720. [Google Scholar] [CrossRef] [Green Version]
- Mayoral, R.; Osborn, O.; McNelis, J.; Johnson, A.M.; Oh, D.Y.; Izquierdo, C.L.; Chung, H.; Li, P.; Traves, P.G.; Bandyopadhyay, G.; et al. Adipocyte SIRT1 knockout promotes PPARgamma activity, adipogenesis and insulin sensitivity in chronic-HFD and obesity. Mol. Metab. 2015, 4, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Wang, L.; Kon, N.; Zhao, W.; Lee, S.; Zhang, Y.; Rosenbaum, M.; Zhao, Y.; Gu, W.; Farmer, S.R.; et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell 2012, 150, 620–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Tong, Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1′s repressive interaction with PPARgamma. Mol. Biol. Cell 2009, 20, 801–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.T.; Chi, K.T.; Lan, Y.W.; Chan, J.C.; Ma, Y.S.; Wei, Y.H. Depletion of Sirt3 leads to the impairment of adipogenic differentiation and insulin resistance via interfering mitochondrial function of adipose-derived human mesenchymal stem cells. Free Radic. Res. 2018, 52, 1398–1415. [Google Scholar] [CrossRef]
- Porter, L.C.; Franczyk, M.P.; Pietka, T.; Yamaguchi, S.; Lin, J.B.; Sasaki, Y.; Verdin, E.; Apte, R.S.; Yoshino, J. NAD(+)-dependent deacetylase SIRT3 in adipocytes is dispensable for maintaining normal adipose tissue mitochondrial function and whole body metabolism. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E520–E530. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Wang, F.; Stieren, E.; Tong, Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem. 2005, 280, 13560–13567. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Li, S.; Wang, X.; Mei, C.; Zan, L. Study of expression analysis of SIRT4 and the coordinate regulation of bovine adipocyte differentiation by SIRT4 and its transcription factors. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Shuai, L.; Zhang, L.N.; Li, B.H.; Tang, C.L.; Wu, L.Y.; Li, J.; Li, J.Y. SIRT5 Regulates Brown Adipocyte Differentiation and Browning of Subcutaneous White Adipose Tissue. Diabetes 2019, 68, 1449–1461. [Google Scholar] [CrossRef]
- Xiong, X.; Zhang, C.; Zhang, Y.; Fan, R.; Qian, X.; Dong, X.C. Fabp4-Cre-mediated Sirt6 deletion impairs adipose tissue function and metabolic homeostasis in mice. J. Endocrinol. 2017, 233, 307–314. [Google Scholar] [CrossRef]
- Chen, Q.; Hao, W.; Xiao, C.; Wang, R.; Xu, X.; Lu, H.; Chen, W.; Deng, C.X. SIRT6 Is Essential for Adipocyte Differentiation by Regulating Mitotic Clonal Expansion. Cell Rep. 2017, 18, 3155–3166. [Google Scholar] [CrossRef]
- Fang, J.; Ianni, A.; Smolka, C.; Vakhrusheva, O.; Nolte, H.; Kruger, M.; Wietelmann, A.; Simonet, N.G.; Adrian-Segarra, J.M.; Vaquero, A.; et al. Sirt7 promotes adipogenesis in the mouse by inhibiting autocatalytic activation of Sirt1. Proc. Natl. Acad. Sci. USA 2017, 114, E8352–E8361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.E.; Ge, K. Transcriptional and epigenetic regulation of PPARgamma expression during adipogenesis. Cell Biosci. 2014, 4, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugii, S.; Evans, R.M. Epigenetic codes of PPARgamma in metabolic disease. FEBS Lett. 2011, 585, 2121–2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Schones, D.E.; Zhao, K. Characterization of human epigenomes. Curr. Opin. Genet. Dev. 2009, 19, 127–134. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, S.A.; Taverna, S.D.; Zhang, Y.; Briggs, S.D.; Li, J.; Eissenberg, J.C.; Allis, C.D.; Khorasanizadeh, S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001, 20, 5232–5241. [Google Scholar] [CrossRef] [Green Version]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Saha, P.K.; Yang, Q.H.; Lee, S.; Park, J.Y.; Suh, Y.; Lee, S.K.; Chan, L.; Roeder, R.G.; Lee, J.W. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 19229–19234. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.E.; Wang, C.; Xu, S.; Cho, Y.W.; Wang, L.; Feng, X.; Baldridge, A.; Sartorelli, V.; Zhuang, L.; Peng, W.; et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife 2013, 2, e01503. [Google Scholar] [CrossRef]
- Jang, Y.; Broun, A.; Wang, C.; Park, Y.K.; Zhuang, L.; Lee, J.E.; Froimchuk, E.; Liu, C.; Ge, K. H3.3K4M destabilizes enhancer H3K4 methyltransferases MLL3/MLL4 and impairs adipose tissue development. Nucleic Acids Res. 2019, 47, 607–620. [Google Scholar] [CrossRef] [Green Version]
- Lai, B.; Lee, J.E.; Jang, Y.; Wang, L.; Peng, W.; Ge, K. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res. 2017, 45, 6388–6403. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.W.; Hong, T.; Hong, S.; Guo, H.; Yu, H.; Kim, D.; Guszczynski, T.; Dressler, G.R.; Copeland, T.D.; Kalkum, M.; et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007, 282, 20395–20406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.E.; Cho, Y.W.; Deng, C.X.; Ge, K. MLL3/MLL4-Associated PAGR1 Regulates Adipogenesis by Controlling Induction of C/EBPbeta and C/EBPdelta. Mol. Cell. Biol. 2020, 40. [Google Scholar] [CrossRef]
- Ohno, H.; Shinoda, K.; Ohyama, K.; Sharp, L.Z.; Kajimura, S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 2013, 504, 163–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Xu, S.; Lee, J.E.; Baldridge, A.; Grullon, S.; Peng, W.; Ge, K. Histone H3K9 methyltransferase G9a represses PPARgamma expression and adipogenesis. EMBO J. 2013, 32, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Nakaki, R.; Inagaki, T.; Yoshida, A.; Kano, Y.; Kimura, H.; Tanaka, T.; Tsutsumi, S.; Nakao, M.; Doi, T.; et al. H3K4/H3K9me3 Bivalent Chromatin Domains Targeted by Lineage-Specific DNA Methylation Pauses Adipocyte Differentiation. Mol. Cell. 2015, 60, 584–596. [Google Scholar] [CrossRef] [Green Version]
- Jing, J.; Li, F.; Zha, L.; Yang, X.; Wu, R.; Wang, S.; Xue, B.; Shi, H. The histone methyltransferase Suv39h regulates 3T3-L1 adipogenesis. Adipocyte 2020, 9, 401–414. [Google Scholar] [CrossRef]
- Wang, L.; Jin, Q.; Lee, J.E.; Su, I.H.; Ge, K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 7317–7322. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, L.; Jang, Y.; Park, Y.K.; Lee, J.E.; Jain, S.; Froimchuk, E.; Broun, A.; Liu, C.; Gavrilova, O.; Ge, K. Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nat. Commun. 2018, 9, 1796. [Google Scholar] [CrossRef] [Green Version]
- Pedrotti, S.; Caccia, R.; Neguembor, M.V.; Garcia-Manteiga, J.M.; Ferri, G.; de Palma, C.; Canu, T.; Giovarelli, M.; Marra, P.; Fiocchi, A.; et al. The Suv420h histone methyltransferases regulate PPAR-gamma and energy expenditure in response to environmental stimuli. Sci. Adv. 2019, 5, 1472. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Zhang, Z.; Rong, W.; Jin, W.; Yan, L.; Jin, W.; Xu, Y.; Cui, X.; Tang, Q.Q.; Pan, D. KMT5c modulates adipocyte thermogenesis by regulating Trp53 expression. Proc. Natl. Acad. Sci. USA 2020, 117, 22413–22422. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, S.E.; Konda, S.; Wu, Q.; Hu, Y.J.; Oslowski, C.M.; Sif, S.; Imbalzano, A.N. Protein arginine methyltransferase 5 (Prmt5) promotes gene expression of peroxisome proliferator-activated receptor gamma2 (PPARgamma2) and its target genes during adipogenesis. Mol. Endocrinol. 2012, 26, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; So, Y.S.; Bae, G.U.; Kim, S.N.; Kim, Y.K. Protein arginine methyltransferase 6 suppresses adipogenic differentiation by repressing peroxisome proliferatoractivated receptor gamma activity. Int. J. Mol. Med. 2019, 43, 2462–2470. [Google Scholar]
- Musri, M.M.; Carmona, M.C.; Hanzu, F.A.; Kaliman, P.; Gomis, R.; Parrizas, M. Histone demethylase LSD1 regulates adipogenesis. J. Biol. Chem. 2010, 285, 30034–30041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duteil, D.; Metzger, E.; Willmann, D.; Karagianni, P.; Friedrichs, N.; Greschik, H.; Gunther, T.; Buettner, R.; Talianidis, I.; Metzger, D.; et al. LSD1 promotes oxidative metabolism of white adipose tissue. Nat. Commun. 2014, 5, 4093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duteil, D.; Tosic, M.; Willmann, D.; Georgiadi, A.; Kanouni, T.; Schule, R. Lsd1 prevents age-programed loss of beige adipocytes. Proc. Natl. Acad. Sci. USA 2017, 114, 5265–5270. [Google Scholar] [CrossRef] [Green Version]
- Takase, R.; Hino, S.; Nagaoka, K.; Anan, K.; Kohrogi, K.; Araki, H.; Hino, Y.; Sakamoto, A.; Nicholson, T.B.; Chen, T.; et al. Lysine-specific demethylase-2 is distinctively involved in brown and beige adipogenic differentiation. FASEB J. 2019, 33, 5300–5311. [Google Scholar] [CrossRef]
- Brier, A.B.; Loft, A.; Madsen, J.G.S.; Rosengren, T.; Nielsen, R.; Schmidt, S.F.; Liu, Z.; Yan, Q.; Gronemeyer, H.; Mandrup, S. The KDM5 family is required for activation of pro-proliferative cell cycle genes during adipocyte differentiation. Nucleic Acids Res. 2017, 45, 1743–1759. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Guo, Y.Y.; Li, B.Y.; Peng, W.Q.; Tang, Q.Q. Histone demethylase KDM5A is transactivated by the transcription factor C/EBPbeta and promotes preadipocyte differentiation by inhibiting Wnt/beta-catenin signaling. J. Biol. Chem. 2019, 294, 9642–9654. [Google Scholar] [CrossRef]
- Varaljai, R.; Islam, A.B.; Beshiri, M.L.; Rehman, J.; Lopez-Bigas, N.; Benevolenskaya, E.V. Increased mitochondrial function downstream from KDM5A histone demethylase rescues differentiation in pRB-deficient cells. Genes Dev. 2015, 29, 1817–1834. [Google Scholar] [CrossRef] [Green Version]
- Tateishi, K.; Okada, Y.; Kallin, E.M.; Zhang, Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 2009, 458, 757–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inagaki, T.; Tachibana, M.; Magoori, K.; Kudo, H.; Tanaka, T.; Okamura, M.; Naito, M.; Kodama, T.; Shinkai, Y.; Sakai, J. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 2009, 14, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Rozqie, R.; Matsumura, Y.; Kawamura, T.; Nakaki, R.; Tsurutani, Y.; Tanimura-Inagaki, K.; Shiono, A.; Magoori, K.; Nakamura, K.; et al. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat. Commun. 2015, 6, 7052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buerger, F.; Muller, S.; Ney, N.; Weiner, J.; Heiker, J.T.; Kallendrusch, S.; Kovacs, P.; Schleinitz, D.; Thiery, J.; Stadler, S.C.; et al. Depletion of Jmjd1c impairs adipogenesis in murine 3T3-L1 cells. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1709–1717. [Google Scholar] [CrossRef]
- Jang, M.K.; Kim, J.H.; Jung, M.H. Histone H3K9 Demethylase JMJD2B Activates Adipogenesis by Regulating H3K9 Methylation on PPARgamma and C/EBPalpha during Adipogenesis. PLoS ONE 2017, 12, e0168185. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Lee, H. Histone demethylase KDM4D cooperates with NFIB and MLL1 complex to regulate adipogenic differentiation of C3H10T1/2 mesenchymal stem cells. Sci. Rep. 2020, 10, 3050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Q.; Wang, Y.; Wang, X.; Yang, J.; Xie, Y.; Zhou, J.; Li, X.; Wang, B. Histone demethylase KDM4A regulates adipogenic and osteogenic differentiation via epigenetic regulation of C/EBPalpha and canonical Wnt signaling. Cell. Mol. Life Sci. 2020, 77, 2407–2421. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, G.; Wang, Y.; Zhou, J.; Yuan, H.; Li, X.; Liu, Y.; Wang, B. Histone demethylase KDM7A reciprocally regulates adipogenic and osteogenic differentiation via regulation of C/EBPalpha and canonical Wnt signalling. J. Cell. Mol. Med. 2019, 23, 2149–2162. [Google Scholar] [CrossRef]
- Okuno, Y.; Ohtake, F.; Igarashi, K.; Kanno, J.; Matsumoto, T.; Takada, I.; Kato, S.; Imai, Y. Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes 2013, 62, 1426–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, D.; Huang, L.; Zhu, L.J.; Zou, T.; Ou, J.; Zhou, W.; Wang, Y.X. Jmjd3-Mediated H3K27me3 Dynamics Orchestrate Brown Fat Development and Regulate White Fat Plasticity. Dev. Cell 2015, 35, 568–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zha, L.; Li, F.; Wu, R.; Artinian, L.; Rehder, V.; Yu, L.; Liang, H.; Xue, B.; Shi, H. The Histone Demethylase UTX Promotes Brown Adipocyte Thermogenic Program Via Coordinated Regulation of H3K27 Demethylation and Acetylation. J. Biol. Chem. 2015, 290, 25151–25163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, S.M.; Jones, P.A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 1979, 17, 771–779. [Google Scholar] [CrossRef]
- Yokomori, N.; Tawata, M.; Onaya, T. DNA demethylation during the differentiation of 3T3-L1 cells affects the expression of the mouse GLUT4 gene. Diabetes 1999, 48, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Kiskinis, E.; Hallberg, M.; Christian, M.; Olofsson, M.; Dilworth, S.M.; White, R.; Parker, M.G. RIP140 directs histone and DNA methylation to silence Ucp1 expression in white adipocytes. EMBO J. 2007, 26, 4831–4840. [Google Scholar] [CrossRef] [PubMed]
- Shore, A.; Karamitri, A.; Kemp, P.; Speakman, J.R.; Lomax, M.A. Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue. Diabetologia 2010, 53, 1164–1173. [Google Scholar] [CrossRef] [Green Version]
- Milagro, F.I.; Campion, J.; Cordero, P.; Goyenechea, E.; Gomez-Uriz, A.M.; Abete, I.; Zulet, M.A.; Martinez, J.A. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J. 2011, 25, 1378–1389. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.C.; Chia, S.Y.; Jin, S.; Han, W.; Ding, C.; Sun, L. Dynamic DNA methylation landscape defines brown and white cell specificity during adipogenesis. Mol. Metab. 2016, 5, 1033–1041. [Google Scholar] [CrossRef]
- Zych, J.; Stimamiglio, M.A.; Senegaglia, A.C.; Brofman, P.R.; Dallagiovanna, B.; Goldenberg, S.; Correa, A. The epigenetic modifiers 5-aza-2′-deoxycytidine and trichostatin A influence adipocyte differentiation in human mesenchymal stem cells. Braz. J. Med. Biol. Res. 2013, 46, 405–416. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, N.; Huang, X.; Xu, J.; Fernandes, J.C.; Dai, K.; Zhang, X. Dexamethasone shifts bone marrow stromal cells from osteoblasts to adipocytes by C/EBPalpha promoter methylation. Cell Death Dis. 2013, 4, e832. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Goldberg, J.; Bremner, J.D.; Vaccarino, V. Global DNA methylation is associated with insulin resistance: A monozygotic twin study. Diabetes 2012, 61, 542–546. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Wu, R.; Shan, W.; Yu, L.; Xue, B.; Shi, H. DNA Methylation Biphasically Regulates 3T3-L1 Preadipocyte Differentiation. Mol. Endocrinol. 2016, 30, 677–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Kang, S. Functional Implications of DNA Methylation in Adipose Biology. Diabetes 2019, 68, 871–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamei, Y.; Suganami, T.; Ehara, T.; Kanai, S.; Hayashi, K.; Yamamoto, Y.; Miura, S.; Ezaki, O.; Okano, M.; Ogawa, Y. Increased expression of DNA methyltransferase 3a in obese adipose tissue: Studies with transgenic mice. Obesity (Silver Spring) 2010, 18, 314–321. [Google Scholar] [CrossRef] [PubMed]
- You, D.; Nilsson, E.; Tenen, D.E.; Lyubetskaya, A.; Lo, J.C.; Jiang, R.; Deng, J.; Dawes, B.A.; Vaag, A.; Ling, C.; et al. Dnmt3a is an epigenetic mediator of adipose insulin resistance. eLife 2017, 6, e30766. [Google Scholar] [CrossRef]
- Barres, R.; Osler, M.E.; Yan, J.; Rune, A.; Fritz, T.; Caidahl, K.; Krook, A.; Zierath, J.R. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009, 10, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Londono Gentile, T.; Lu, C.; Lodato, P.M.; Tse, S.; Olejniczak, S.H.; Witze, E.S.; Thompson, C.B.; Wellen, K.E. DNMT1 is regulated by ATP-citrate lyase and maintains methylation patterns during adipocyte differentiation. Mol. Cell. Biol. 2013, 33, 3864–3878. [Google Scholar] [CrossRef] [Green Version]
- Li, J.J.F.; Movahed, M.; Cui, X.; Cao, Q.; Wu, R.; Chen, Z.; Yu, L.; Pan, Y.; Shi, H.; Xue, B.; et al. Epigenetic Interaction between UTX and DNMT1 Regulates Diet-Induced Myogenic Remodeling in Brown Fat. bioRxiv 2020. [Google Scholar] [CrossRef]
- Damal Villivalam, S.; You, D.; Kim, J.; Lim, H.W.; Xiao, H.; Zushin, P.H.; Oguri, Y.; Amin, P.; Kang, S. TET1 is a beige adipocyte-selective epigenetic suppressor of thermogenesis. Nat. Commun. 2020, 11, 4313. [Google Scholar] [CrossRef]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [Green Version]
- White, U.A.; Stephens, J.M. Transcriptional factors that promote formation of white adipose tissue. Mol. Cell. Endocrinol. 2010, 318, 10–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarjeant, K.; Stephens, J.M. Adipogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tontonoz, P.; Hu, E.; Spiegelman, B.M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994, 79, 1147–1156. [Google Scholar] [CrossRef]
- Lee, J.E.; Schmidt, H.; Lai, B.; Ge, K. Transcriptional and Epigenomic Regulation of Adipogenesis. Mol. Cell. Biol. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J., Jr.; Liu, X.S.; et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; Walkey, C.J.; Puigserver, P.; Spiegelman, B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000, 14, 1293–1307. [Google Scholar]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Conroe, H.M.; Estall, J.; Kajimura, S.; Frontini, A.; Ishibashi, J.; Cohen, P.; Cinti, S.; Spiegelman, B.M. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Investig. 2011, 121, 96–105. [Google Scholar] [CrossRef] [Green Version]
- Sambeat, A.; Gulyaeva, O.; Dempersmier, J.; Sul, H.S. Epigenetic Regulation of the Thermogenic Adipose Program. Trends Endocrinol. Metab. 2017, 28, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Zhang, Q.; Liao, C.; Han, W.; Xu, F. Epigenomic Control of Thermogenic Adipocyte Differentiation and Function. Int. J. Mol. Sci. 2018, 19, 1793. [Google Scholar] [CrossRef] [Green Version]
- Villanueva, C.J.; Waki, H.; Godio, C.; Nielsen, R.; Chou, W.L.; Vargas, L.; Wroblewski, K.; Schmedt, C.; Chao, L.C.; Boyadjian, R.; et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 2011, 13, 413–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villanueva, C.J.; Vergnes, L.; Wang, J.; Drew, B.G.; Hong, C.; Tu, Y.; Hu, Y.; Peng, X.; Xu, F.; Saez, E.; et al. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARgamma specifies lipid storage versus thermogenic gene programs. Cell Metab. 2013, 17, 423–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearson, S.; Loft, A.; Rajbhandari, P.; Simcox, J.; Lee, S.; Tontonoz, P.; Mandrup, S.; Villanueva, C.J. Loss of TLE3 promotes the mitochondrial program in beige adipocytes and improves glucose metabolism. Genes Dev. 2019, 33, 747–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.K.; Arany, Z.; Seale, P.; Mepani, R.J.; Ye, L.; Conroe, H.M.; Roby, Y.A.; Kulaga, H.; Reed, R.R.; Spiegelman, B.M. Transcriptional control of preadipocyte determination by Zfp423. Nature 2010, 464, 619–623. [Google Scholar] [CrossRef] [Green Version]
- Shao, M.; Ishibashi, J.; Kusminski, C.M.; Wang, Q.A.; Hepler, C.; Vishvanath, L.; MacPherson, K.A.; Spurgin, S.B.; Sun, K.; Holland, W.L.; et al. Zfp423 Maintains White Adipocyte Identity through Suppression of the Beige Cell Thermogenic Gene Program. Cell Metab. 2016, 23, 1167–1184. [Google Scholar] [CrossRef] [Green Version]
- Kita, M.; Nakae, J.; Kawano, Y.; Asahara, H.; Takemori, H.; Okado, H.; Itoh, H. Zfp238 Regulates the Thermogenic Program in Cooperation with Foxo1. iScience 2019, 12, 87–101. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Maekawa, T.; Yoshida, K.; Furuse, T.; Kaneda, H.; Wakana, S.; Ishii, S. ATF7 ablation prevents diet-induced obesity and insulin resistance. Biochem. Biophys. Res. Commun. 2016, 478, 696–702. [Google Scholar] [CrossRef]
- Liu, Y.; Maekawa, T.; Yoshida, K.; Muratani, M.; Chatton, B.; Ishii, S. The Transcription Factor ATF7 Controls Adipocyte Differentiation and Thermogenic Gene Programming. iScience 2019, 13, 98–112. [Google Scholar] [CrossRef] [Green Version]
- Patil, M.; Sharma, B.K.; Elattar, S.; Chang, J.; Kapil, S.; Yuan, J.; Satyanarayana, A. Id1 Promotes Obesity by Suppressing Brown Adipose Thermogenesis and White Adipose Browning. Diabetes 2017, 66, 1611–1625. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, M.; Dahlman, I.; Ryden, M.; Nordstrom, E.A.; Gustafsson, J.A.; Arner, P.; Dahlman-Wright, K. Oestrogen receptor alpha gene expression levels are reduced in obese compared to normal weight females. Int. J. Obes. 2007, 31, 900–907. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Moore, T.M.; Drew, B.G.; Ribas, V.; Wanagat, J.; Civelek, M.; Segawa, M.; Wolf, D.M.; Norheim, F.; Seldin, M.M.; et al. Estrogen receptor alpha controls metabolism in white and brown adipocytes by regulating Polg1 and mitochondrial remodeling. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Santos, R.S.; Frank, A.P.; Fatima, L.A.; Palmer, B.F.; Oz, O.K.; Clegg, D.J. Activation of estrogen receptor alpha induces beiging of adipocytes. Mol. Metab. 2018, 18, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.F.; Su, W.; Dai, Y.B.; Wu, W.F.; Huang, B.; Barros, R.P.; Nguyen, H.; Maneix, L.; Guan, Y.F.; Warner, M.; et al. An ERbeta agonist induces browning of subcutaneous abdominal fat pad in obese female mice. Sci. Rep. 2016, 6, 38579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reggiani, L.; Raciti, D.; Airik, R.; Kispert, A.; Brandli, A.W. The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev. 2007, 21, 2358–2370. [Google Scholar] [CrossRef] [Green Version]
- Smemo, S.; Tena, J.J.; Kim, K.H.; Gamazon, E.R.; Sakabe, N.J.; Gomez-Marin, C.; Aneas, I.; Credidio, F.L.; Sobreira, D.R.; Wasserman, N.F.; et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 2014, 507, 371–375. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Schill, R.L.; O’Donnell, M.; Xu, K.; Bagchi, D.P.; MacDougald, O.A.; Koenig, R.J.; Xu, B. The transcription factor NKX1-2 promotes adipogenesis and may contribute to a balance between adipocyte and osteoblast differentiation. J. Biol. Chem. 2019, 294, 18408–18420. [Google Scholar] [CrossRef]
- Xu, B.; O’Donnell, M.; O’Donnell, J.; Yu, J.; Zhang, Y.; Sartor, M.A.; Koenig, R.J. Adipogenic Differentiation of Thyroid Cancer Cells Through the Pax8-PPARgamma Fusion Protein Is Regulated by Thyroid Transcription Factor 1 (TTF-1). J. Biol. Chem. 2016, 291, 19274–19286. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Pan, D.; Chen, Q.; Zhu, L.J.; Ou, J.; Wabitsch, M.; Wang, Y.X. Transcription factor Hlx controls a systematic switch from white to brown fat through Prdm16-mediated co-activation. Nat. Commun. 2017, 8, 68. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Zhou, Y.; Yang, Y.; Peng, J.; Song, T.; Xu, T.; Wei, H.; Jiang, S.; Peng, J. Identification of zinc finger protein Bcl6 as a novel regulator of early adipose commitment. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Senagolage, M.D.; Sommars, M.A.; Ramachandran, K.; Futtner, C.R.; Omura, Y.; Allred, A.L.; Wang, J.; Yang, C.; Procissi, D.; Evans, R.M.; et al. Loss of Transcriptional Repression by BCL6 Confers Insulin Sensitivity in the Setting of Obesity. Cell Rep. 2018, 25, 3283–3298. [Google Scholar] [CrossRef] [Green Version]
- Kutyavin, V.I.; Chawla, A. BCL6 regulates brown adipocyte dormancy to maintain thermogenic reserve and fitness. Proc. Natl. Acad. Sci. USA 2019, 116, 17071–17080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.B.; Kong, M.; Kim, T.M.; Suh, Y.H.; Kim, W.H.; Lim, J.H.; Song, J.H.; Jung, M.H. NFATc4 and ATF3 negatively regulate adiponectin gene expression in 3T3-L1 adipocytes. Diabetes 2006, 55, 1342–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, M.K.; Kim, C.H.; Seong, J.K.; Jung, M.H. ATF3 inhibits adipocyte differentiation of 3T3-L1 cells. Biochem. Biophys. Res. Commun. 2012, 421, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.K.; Jung, M.H. ATF3 represses PPARgamma expression and inhibits adipocyte differentiation. Biochem. Biophys. Res. Commun. 2014, 454, 58–64. [Google Scholar] [CrossRef]
- Jang, M.K.; Son, Y.; Jung, M.H. ATF3 plays a role in adipocyte hypoxia-mediated mitochondria dysfunction in obesity. Biochem. Biophys. Res. Commun. 2013, 431, 421–427. [Google Scholar] [CrossRef]
- Cheng, C.F.; Ku, H.C.; Cheng, J.J.; Chao, S.W.; Li, H.F.; Lai, P.F.; Chang, C.C.; Don, M.J.; Chen, H.H.; Lin, H. Adipocyte browning and resistance to obesity in mice is induced by expression of ATF3. Commun. Biol. 2019, 2, 389. [Google Scholar] [CrossRef]
- Milet, C.; Bleher, M.; Allbright, K.; Orgeur, M.; Coulpier, F.; Duprez, D.; Havis, E. Egr1 deficiency induces browning of inguinal subcutaneous white adipose tissue in mice. Sci. Rep. 2017, 7, 16153. [Google Scholar] [CrossRef] [Green Version]
- Bleher, M.; Meshko, B.; Cacciapuoti, I.; Gergondey, R.; Kovacs, Y.; Duprez, D.; L’Honore, A.; Havis, E. Egr1 loss-of-function promotes beige adipocyte differentiation and activation specifically in inguinal subcutaneous white adipose tissue. Sci. Rep. 2020, 10, 15842. [Google Scholar] [CrossRef]
- Grabowska, A.; Pakula, B.; Pancir, J. Excited states of six-membered N-heterocycles. Fluorescence, phosphorescence and acid-base equilibria of five mono- and diazaphenanthrenes in the lowest excited (pi,pi) states. Photochem. Photobiol. 1969, 10, 415–425. [Google Scholar] [CrossRef]
- Ng, R.; Hussain, N.A.; Zhang, Q.; Chang, C.; Li, H.; Fu, Y.; Cao, L.; Han, W.; Stunkel, W.; Xu, F. miRNA-32 Drives Brown Fat Thermogenesis and Trans-activates Subcutaneous White Fat Browning in Mice. Cell Rep. 2017, 19, 1229–1246. [Google Scholar] [CrossRef] [Green Version]
- Virtue, A.T.; McCright, S.J.; Wright, J.M.; Jimenez, M.T.; Mowel, W.K.; Kotzin, J.J.; Joannas, L.; Basavappa, M.G.; Spencer, S.P.; Clark, M.L.; et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Acharya, A.; Berry, D.C.; Zhang, H.; Jiang, Y.; Jones, B.T.; Hammer, R.E.; Graff, J.M.; Mendell, J.T. miR-26 suppresses adipocyte progenitor differentiation and fat production by targeting Fbxl19. Genes Dev. 2019, 33, 1367–1380. [Google Scholar] [CrossRef] [Green Version]
- Arias, N.; Aguirre, L.; Fernandez-Quintela, A.; Gonzalez, M.; Lasa, A.; Miranda, J.; Macarulla, M.T.; Portillo, M.P. MicroRNAs involved in the browning process of adipocytes. J. Physiol. Biochem. 2016, 72, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Scheideler, M. MicroRNAs in adipocyte formation and obesity. Best Pract. Res. Clin. Endocrinol. Metab. 2016, 30, 653–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trajkovski, M.; Ahmed, K.; Esau, C.C.; Stoffel, M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat. Cell Biol. 2012, 14, 1330–1335. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Trajkovski, M. MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism 2014, 63, 272–282. [Google Scholar] [CrossRef]
- Chou, C.F.; Lin, Y.Y.; Wang, H.K.; Zhu, X.; Giovarelli, M.; Briata, P.; Gherzi, R.; Garvey, W.T.; Chen, C.Y. KSRP ablation enhances brown fat gene program in white adipose tissue through reduced miR-150 expression. Diabetes 2014, 63, 2949–2961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, T.; Seok, S.; Choi, S.; Huang, Z.; Suino-Powell, K.; Xu, H.E.; Kemper, B.; Kemper, J.K. MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol. Cell. Biol. 2014, 34, 4130–4142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.; Nakagami, H.; Rodriguez-Araujo, G.; Nimura, K.; Kaneda, Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2012, 10, e1001314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Siegel, F.; Kipschull, S.; Haas, B.; Frohlich, H.; Meister, G.; Pfeifer, A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat. Commun. 2013, 4, 1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Guan, M.; Townsend, K.L.; Huang, T.L.; An, D.; Yan, X.; Xue, R.; Schulz, T.J.; Winnay, J.; Mori, M.; et al. MicroRNA-455 regulates brown adipogenesis via a novel HIF1an-AMPK-PGC1alpha signaling network. EMBO Rep. 2015, 16, 1378–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, F.; Wang, M.; Xiao, T.; Yin, B.; He, L.; Meng, W.; Dong, M.; Liu, F. miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes 2015, 64, 2056–2068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giroud, M.; Karbiener, M.; Pisani, D.F.; Ghandour, R.A.; Beranger, G.E.; Niemi, T.; Taittonen, M.; Nuutila, P.; Virtanen, K.A.; Langin, D.; et al. Let-7i-5p represses brite adipocyte function in mice and humans. Sci. Rep. 2016, 6, 28613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giroud, M.; Pisani, D.F.; Karbiener, M.; Barquissau, V.; Ghandour, R.A.; Tews, D.; Fischer-Posovszky, P.; Chambard, J.C.; Knippschild, U.; Niemi, T.; et al. miR-125b affects mitochondrial biogenesis and impairs brite adipocyte formation and function. Mol. Metab. 2016, 5, 615–625. [Google Scholar] [CrossRef]

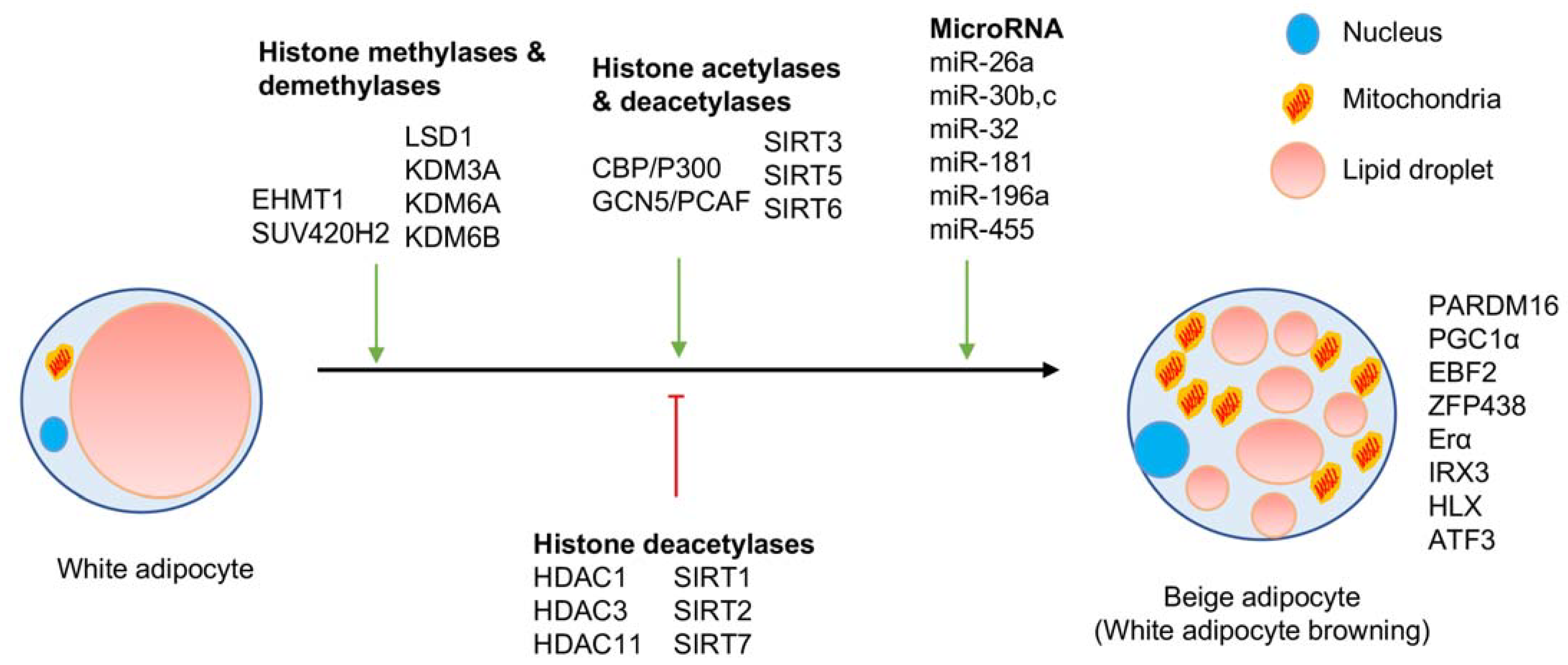
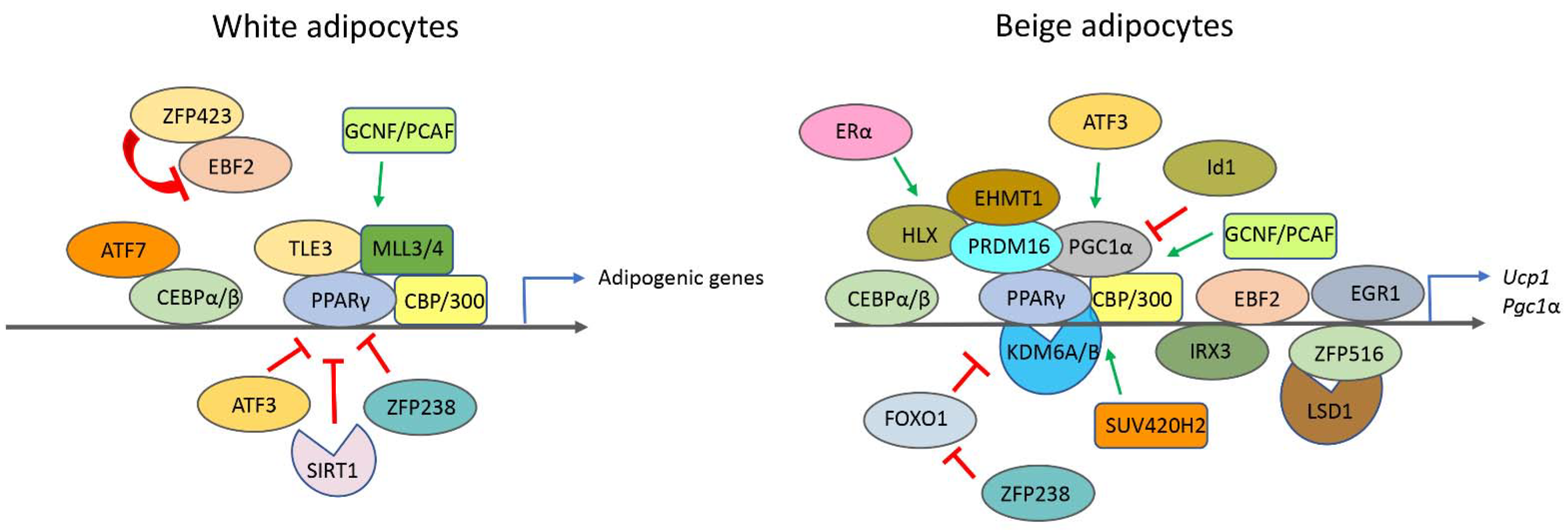
| Histone Modification | Effector | Epigenetic Mark | Role in Adipogenesis or White Browning | Reference |
|---|---|---|---|---|
| Histone acetylation | CBP and P300 | H3K27ac | Promote adipogenesis | [21] |
| H3K27ac | Promotes white adipocyte browning | [12] | ||
| GCN5 and PCAF | H3K9ac | Promotes WAT browning and brown preadipocyte differentiation | [24,25] | |
| Histone deacetylation | HDAC1 and HDAC2 | H3K27ac | Promotes adipogenesis | [33] |
| HDAC3 | H3K27ac | Promotes adipogenesis and inhibits WAT browning | [31] | |
| HDAC9 | H3K27ac | Inhibits adipogenesis | [35] | |
| HDAC11 | H3K27ac | Inhibits white adipocyte browning | [32] | |
| SIRT1 | H3K9ac, H4K16ac | Inhibits adipogenesis but promotes white adipocyte browning | [41,42,43] | |
| SIRT2 | H3K9ac | Inhibits adipogenesis and white adipocyte browning | [44] | |
| SIRT3 | H3K9ac | Promotes white adipocyte browning | [47] | |
| SIRT5 | H3K9ac | Inhibits white adipocyte browning | [49] | |
| SIRT6 | H3K9ac | Promotes adipogenesis | [51] | |
| SIRT7 | H3K9ac, H4K16ac | Inhibits adipogenesis | [52] | |
| Histone Methylation | MLL3 | H3K4me3 | Promotes adipogenesis | [59] |
| MLL4 | H3K4me3 | Promotes adipogenesis | [60,61] | |
| EHMT1 | H3K9me2/3 | Promotes white adipocyte browning | [65] | |
| G9A | H3K9me2 | Inhibits adipogenesis | [66] | |
| SETDB1 | H3K9me3 | Inhibits adipogenesis | [67] | |
| SUV39H1 SUV39H2 | >H3K9me2/me3 | Promotes adipogenesis | [68] | |
| EZH2 | H3K27me3 | Promotes adipogenesis | [69] | |
| NSD2 | H3K36me3 | Promotes adipogenesis | [70] | |
| >SUV420H2 | H3K20me3 | Promotes white adipocyte browning | [72] | |
| PRMT5 | H3R8me2 | Promotes adipogenesis | [73] | |
| PRMT6 | H3R2me2 | Inhibits adipogenesis | [74] | |
| Histone Demethylation | KDM1A | H3K4me2 | Promotes adipogenesis and white adipocyte browning | [77] |
| KDM1B | H3K4me2 | Promotes adipogenesis | [78] | |
| KDM5A | H3K4me3 | Promotes adipogenesis | [80] | |
| KDM3A | H3K9me | Promotes white adipocyte browning | [82,83,84] | |
| KDM3C | H3K9me | Promotes adipogenesis | [85] | |
| KDM4B | H3K9me | Promotes adipogenesis | [86] | |
| KDM4D | H3K9me3 | Promotes adipogenesis | [87] | |
| KDM4A | H3K9me3 | Promotes adipogenesis | [88] | |
| KDM7A | H3K9me2, H3K27me2 | Promotes adipogenesis | [89] | |
| KDM7C | H3K9me2 | Promotes adipogenesis | [90] | |
| KDM6B | H3k27me3 | Promotes white adipocyte browning | [91] | |
| KDM6A | H3k27me3 | Promotes white adipocyte browning | [92] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanduri, R. Epigenetic Regulators of White Adipocyte Browning. Epigenomes 2021, 5, 3. https://0-doi-org.brum.beds.ac.uk/10.3390/epigenomes5010003
Nanduri R. Epigenetic Regulators of White Adipocyte Browning. Epigenomes. 2021; 5(1):3. https://0-doi-org.brum.beds.ac.uk/10.3390/epigenomes5010003
Chicago/Turabian StyleNanduri, Ravikanth. 2021. "Epigenetic Regulators of White Adipocyte Browning" Epigenomes 5, no. 1: 3. https://0-doi-org.brum.beds.ac.uk/10.3390/epigenomes5010003





