Atypical Non-H2S-Producing Monophasic Salmonella Typhimurium ST3478 Strains from Chicken Meat at Processing Stage Are Adapted to Diverse Stresses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Strategy in the Slaughterhouse and Processing Plant
2.2. Detection and Characterization of Salmonella by a Cultural Approach
2.3. Detection of Salmonella by a Molecular Approach
2.4. Phenotypic and Genotypic Characterization of Salmonella Isolates Recovered from Positive Samples
2.5. Whole-Genome Sequencing (WGS) for Characterization of Salmonella Isolates
2.6. Comparative Genomic Analysis of Salmonella ST3478 Isolates
2.7. Nucleotide Sequence Accession Numbers
3. Results and Discussion
3.1. Low Salmonella Occurrence among Raw Chicken Meat by Conventional and Molecular Methods
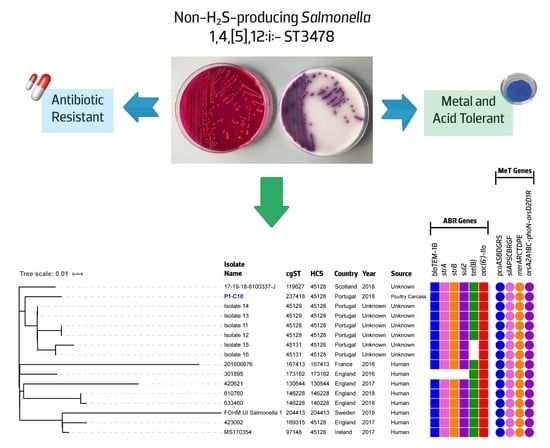
3.2. Atypical Non-H2S-Producing S. 1,4,[5],12:i:- ST3478 with the Ability to Tolerate Diverse Food Chain Stresses
3.3. Comparative Genomics Reveals an Ongoing Emergence of a Non-H2S-Producing S. 1,4,[5],12:i:-/ST3478 Clonal Lineage
3.3.1. Non-H2S-Producing Phenotype Conferred by a Nonsense Mutation in the phsA Thiosulfate Reductase Gene is Increasingly Reported
3.3.2. Comparative Genomic of Global S. 1,4,[5],12:i:- ST3478 Showed Common Adaptive Features and Genetic Backgrounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, 5926. [Google Scholar]
- The European Parliament; Council of the European Union. Regulation (EC) No 2160/2003 of the European Parliament and of the Council of 17 November 2003 on the Control of Salmonella and Other Specified Food-Borne Zoonotic Agents; EU: Brussels, Belgium, 2003. [Google Scholar]
- The European Commission. Commission Regulation (EU) No 1086/2011 of 27 October 2011 Amending Annex II to Regulation (EC) No 2160/2003 of the European Parliament and of the Council and Annex I to Commission Regulation (EC) No 2073/2005 as Regards Salmonella in Fresh Poultry Meat; The European Commission: Brussels, Belgium, 2011. [Google Scholar]
- The European Commission. Commission Regulation (EU) No 200/2012 of 8 March 2012 Concerning a Union Target for the Reduction of Salmonella Enteritidis and Salmonella Typhimurium in Flocks of Broilers, as Provided for in Regulation (EC) No 2160/2003 of the European Parliament and of the Council; The European Commission: Brussels, Belgium, 2012. [Google Scholar]
- Lin, D.; Yan, M.; Lin, S.; Chen, S. Increasing prevalence of hydrogen sulfide negative Salmonella in retail meats. Food Microbiol. 2014, 43, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.; Mourão, J.; Silveira, L.; Saraiva, M.; Correia, C.B.; Maçãs, A.P.; Peixe, L.; Antunes, P. Imported poultry meat as a source of extended-spectrum cephalosporin-resistant CMY-2-producing Salmonella Heidelberg and Salmonella Minnesota in the European Union, 2014–2015. Int. J. Antimicrob. Agents 2018, 51, 151–154. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019, 17, e05598. [Google Scholar]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Cesare, A.D.; Herman, L.; Hilbert, F.; Lindqvist, R.; et al. Salmonella control in poultry flocks and its public health impact. EFSA J. 2019, 17, e05596. [Google Scholar]
- European Medicines Agency (EMA). Updated Advice on the Use of Colistin Products in Animals within the European Union: Development of Resistance and Possible Impact on Human and Animal Health; European Medicines Agency (EMA): Amsterdam, The Netherlands, 2016. [Google Scholar]
- European Medicines Agency (EMA); European Food Safety Authority (EFSA). Joint Scientific Opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA). EFSA J. 2017, 15, 4666.
- International Organization for Standardization (ISO). ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp.; International Organization for Standardization (ISO): Geneva, Switzerland, 2017. [Google Scholar]
- Pritchett, L.C.; Konkel, M.E.; Gay, J.M.; Besser, T.E. Identification of DT104 and U302 phage types among Salmonella enterica serotype typhimurium isolates by PCR. J. Clin. Microbiol. 2000, 38, 3484–3488. [Google Scholar] [CrossRef] [Green Version]
- Héritier, C.; Poirel, L.; Aubert, D.; Nordmann, P. Genetic and Functional Analysis of the Chromosome-Encoded Carbapenem-Hydrolyzing Oxacillinase OXA-40 of Acinetobacter baumannii. AAC 2003, 47, 268–273. [Google Scholar] [CrossRef] [Green Version]
- Tennant, S.M.; Diallo, S.; Levy, H.; Livio, S.; Sow, S.O.; Tapia, M.; Fields, P.I.; Mikoleit, M.; Tamboura, B.; Kotloff, K.L.; et al. Identification by PCR of Non-typhoidal Salmonella enterica Serovars Associated with Invasive Infections among Febrile Patients in Mali. PLoS Negl. Trop. Dis. 2010, 4, e621. [Google Scholar] [CrossRef] [Green Version]
- Mourão, J.; Novais, C.; Machado, J.; Peixe, L.; Antunes, P. Metal tolerance in emerging clinically relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i: – clones circulating in Europe. Int. J. Antimicrob. Agents 2015, 45, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Mourão, J.; Rebelo, A.; Ribeiro, S.; Peixe, L.; Novais, C.; Antunes, P. Tolerance to arsenic contaminant among multidrug-resistant and copper-tolerant Salmonella successful clones is associated with diverse operons and genetic contexts. Environ. Microbiol. 2020, 22, 2829–2842. [Google Scholar] [CrossRef] [PubMed]
- Mourão, J.; Marçal, S.; Ramos, P.; Campos, J.; Machado, J.; Peixe, L.; Novais, C.; Antunes, P. Tolerance to multiple metal stressors in emerging non-typhoidal MDR Salmonella serotypes: A relevant role for copper in anaerobic conditions. J. Antimicrob. Chemother. 2016, 71, 2147–2157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Committee of Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. 2020. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf (accessed on 12th June 2020).
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI Document M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- European Committee of Antimicrobial Susceptibility Testing (EUCAST). Recommendations for MIC determination of colistin (polymyxin E) As recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. 2016. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf (accessed on 12th June 2020).
- International Organization for Standardization (ISO). ISO 20776-1:2006: Clinical Laboratory Testing and in Vitro Diagnostic Test Systems—Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 1: Reference Method for Testing the In Vitro Activity of Antimicrobial Agents against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases; International Organization for Standardization (ISO): Geneva, Switzerland, 2006. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline; CLSI Document M26-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1999. [Google Scholar]
- El-Azizi, M.; Farag, N.; Khardori, N. Efficacy of selected biocides in the decontamination of common nosocomial bacterial pathogens in biofilm and planktonic forms. Comp. Immunol. Microbiol. Infect. Dis. 2016, 47, 60–71. [Google Scholar] [CrossRef]
- Lange’s Handbook of Chemistry, 13td ed.; Dean, J.A.; Lange, N.A. (Eds.) McGraw-Hill: New York, NY, USA, 1985; ISBN 9780070161924. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Zhou, Z.; Sergeant, M.J.; Achtman, M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018, 14, e1007261. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.J.; Nash, J.H.E.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; Achtman, M.; Brown, D.; Chattaway, M.; Dallman, T.; Delahay, R.; Kornschober, C.; et al. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zheng, Q.; Lin, J.; Yuk, H.-G.; Guo, L. Immuno- and nucleic acid-based current technique for Salmonella detection in food. Eur. Food Res. Technol. 2020, 246, 373–395. [Google Scholar] [CrossRef]
- Bell, R.L.; Jarvis, K.G.; Ottesen, A.R.; McFarland, M.A.; Brown, E.W. Recent and emerging innovations in Salmonella detection: A food and environmental perspective. Microb. Biotechnol. 2016, 9, 279–292. [Google Scholar] [CrossRef]
- Silveira, L.; Pista, Â.; Machado, J. Caracterização fenotípica de isolados de Salmonella enterica recebidos no INSA entre 2014 e 2017. Boletim Epidemiológico Observações 2018, 7, 49–51. [Google Scholar]
- Lin, J.; Lee, I.S.; Frey, J.; Slonczewski, J.L.; Foster, J.W. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 1995, 177, 4097–4104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bearson, S.; Bearson, B.; Foster, J.W. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 1997, 147, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.P.; Kendall, P.A.; Sofos, J.N. Modeling the boundaries of growth of Salmonella Typhimurium in broth as a function of temperature, water activity, and pH. J. Food Prot. 2004, 67, 53–59. [Google Scholar] [CrossRef]
- Hu, S.; Yu, Y.; Zhou, D.; Li, R.; Xiao, X.; Wu, H. Global transcriptomic acid tolerance response in Salmonella Enteritidis. LWT 2018, 92, 330–338. [Google Scholar] [CrossRef]
- Joerger, R.D.; Sartori, C.; Frye, J.G.; Turpin, J.B.; Schmidt, C.; McClelland, M.; Porwollik, S. Gene expression analysis of Salmonella enterica Enteritidis Nal R and Salmonella enterica Kentucky 3795 exposed to hcl and acetic acid in rich medium. Foodborne Pathog. Dis. 2012, 9, 331–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lianou, A.; Nychas, G.-J.E.; Koutsoumanis, K.P. Variability in the adaptive acid tolerance response phenotype of Salmonella enterica strains. Food Microbiol. 2017, 62, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Joerger, R.D.; Sartori, C.A.; Kniel, K.E. Comparison of genetic and physiological properties of Salmonella enterica isolates from chickens reveals one major difference between Serovar Kentucky and Other Serovars: Response to acid. Foodborne Pathog. Dis. 2009, 6, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Dubois-Brissonnet, F. Adaptation of Salmonella to Antimicrobials in Food-Processing Environments. In Salmonella—Distribution, Adaptation, Control Measures and Molecular Technologies; Annous, B., Ed.; InTech: London, UK, 2012; ISBN 9789535106616. [Google Scholar]
- Alonso-Hernando, A.; Alonso-Calleja, C.; Capita, R. Effects of exposure to poultry chemical decontaminants on the membrane fluidity of Listeria monocytogenes and Salmonella enterica strains. Int. J. Food Microbiol. 2010, 137, 130–136. [Google Scholar] [CrossRef]
- Humayoun, S.B.; Hiott, L.M.; Gupta, S.K.; Barrett, J.B.; Woodley, T.A.; Johnston, J.J.; Jackson, C.R.; Frye, J.G. An assay for determining the susceptibility of Salmonella isolates to commercial and household biocides. PLoS ONE 2018, 13, e0209072. [Google Scholar] [CrossRef] [Green Version]
- Micciche, A.C.; Feye, K.M.; Rubinelli, P.M.; Lee, J.A.; Knueven, C.J.; Ricke, S.C. Comparison of acid sanitizers on Salmonella Typhimurium inoculated commercial poultry processing reuse water. Front. Sustain. Food Syst. 2019, 2, 90. [Google Scholar] [CrossRef] [Green Version]
- Jolivet-Gougeon, A.; Sauvager, F.; Bonnaure-Mallet, M.; Colwell, R.R.; Cormier, M. Virulence of viable but nonculturable S. Typhimurium LT2 after peracetic acid treatment. Int. J. Food Microbiol. 2006, 112, 147–152. [Google Scholar] [CrossRef]
- Bauermeister, L.J.; Bowers, J.W.J.; Townsend, J.C.; McKee, S.R. The microbial and quality properties of poultry carcasses treated with peracetic acid as an Antimicrobial Treatment. Poult. Sci. 2008, 87, 2390–2398. [Google Scholar] [CrossRef]
- Mathew, E.N.; Muyyarikkandy, M.S.; Bedell, C.; Amalaradjou, M.A. Efficacy of Chlorine, Chlorine Dioxide, and Peroxyacetic Acid in Reducing Salmonella Contamination in wash water and on mangoes under simulated mango packinghouse washing operations. Front. Sustain. Food Syst. 2018, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Lelieveld, H.; Holah, J.; Napper, D. Hygiene in Food Processing, 2nd ed.; Woodhead Publishing: Cambridge, UK, 2014; ISBN 9780857094292. [Google Scholar]
- European Chemicals Agency (ECHA); Biocidal Products Committee (BPC). Opinion on the application for approval of the active substance: Paracetic Acid—Product type: 4. 2015. Available online: https://echa.europa.eu/documents/10162/24380804/7864_AS-APP_Peracetic+acid_PT4_Final+opinion.pdf/f077147e-e152-fc24-4f46-0b971a0fb25a (accessed on 12th June 2020).
- European Chemicals Agency (ECHA); Biocidal Products Committee (BPC). Regulation (EU) No 528/2012 concerning the making available on the market and use of biocidal products – Evaluation of active substances, Assessment Report of Peracetic Acid (Product Type 1-6). 2015. Available online: https://echa.europa.eu/documents/10162/24380810/8376_1340-04_Assessment_Report.pdf (accessed on 12th June 2020).
- Cadena, M.; Kelman, T.; Marco, M.L.; Pitesky, M. Understanding Antimicrobial Resistance (AMR) Profiles of Salmonella Biofilm and Planktonic Bacteria Challenged with Disinfectants Commonly Used During Poultry Processing. Foods 2019, 8, 275. [Google Scholar] [CrossRef] [Green Version]
- Jourdan-da Silva, N.; Fabre, L.; Robinson, E.; Fournet, N.; Nisavanh, A.; Bruyand, M.; Mailles, A.; Serre, E.; Ravel, M.; Guibert, V.; et al. Ongoing nationwide outbreak of Salmonella Agona associated with internationally distributed infant milk products, France, December 2017. Eurosurveillance 2018, 23, 17–00852. [Google Scholar] [CrossRef] [PubMed]
- Colombe, S.; Jernberg, C.; Löf, E.; Angervall, A.L.; Mellström-Dahlgren, H.; Dotevall, L.; Bengnér, M.; Hall, I.; Sundqvist, L.; Kühlmann-Berenzon, S.; et al. Outbreak of unusual H2S-negative monophasic Salmonella Typhimurium strain likely associated with small tomatoes, Sweden, August to October 2019. Eurosurveillance 2019, 24, 1900643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakano, C.; Kuroda, M.; Sekizuka, T.; Ishioka, T.; Morita, Y.; Ryo, A.; Tsukagoshi, H.; Kawai, Y.; Inoue, N.; Takada, H.; et al. Genetic Analysis of Non-Hydrogen Sulfide-Producing Salmonella enterica Serovar Typhimurium and S. enterica Serovar Infantis Isolates in Japan. J. Clin. Microbiol. 2013, 51, 328–330. [Google Scholar] [PubMed] [Green Version]
- Albert, M.J.; Al Obaid, K.; Alfouzan, W.; Sheikh, A.R.; Udo, E.; Izumiya, H.; Bulach, D.M.; Seemann, T. Isolation of Salmonella enterica Serovar Kentucky Strain ST 198 and Its H2S-Negative Variant from a Patient: Implications for Diagnosis. J. Clin. Microbiol. 2014, 52, 4090–4093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, S.; Xie, J.; Liu, N.; Li, P.; Xu, X.; Li, H.; Sun, J.; Wang, J.; Liang, B.; Yang, C.; et al. Emergence and prevalence of Non-H2S-Producing Salmonella enterica serovar senftenberg isolates belonging to novel sequence type 1751 in China. J. Clin. Microbiol. 2014, 52, 2557–2565. [Google Scholar] [CrossRef] [Green Version]
- Heinzinger, N.K.; Fujimoto, S.Y.; Clark, M.A.; Moreno, M.S.; Barrett, E.L. Sequence analysis of the phs operon in Salmonella typhimurium and the contribution of thiosulfate reduction to anaerobic energy metabolism. J. Bacteriol. 1995, 177, 2813–2820. [Google Scholar] [CrossRef] [Green Version]
- Rohmer, L.; Hocquet, D.; Miller, S.I. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011, 19, 341–348. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Xu, X.; Xie, J.; Yi, S.; Wang, J.; Yang, X.; Yang, C.; Liang, B.; Ma, Q.; Li, H.; et al. Molecular characterization of Salmonella enterica Serovar Aberdeen Negative for H2S production in China. PLoS ONE 2016, 11, e0161352. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Yi, S.; Zhu, J.; Li, P.; Liang, B.; Li, H.; Yang, X.; Wang, L.; Hao, R.; Jia, L.; et al. Antimicrobial resistance and molecular investigation of H2S-Negative Salmonella enterica subsp. enterica serovar Choleraesuis Isolates in China. PLoS ONE 2015, 10, e0139115. [Google Scholar]
- Xie, J.; Wu, F.; Xu, X.; Yang, X.; Zhao, R.; Ma, Q.; Li, P.; Wang, L.; Hao, R.; Jia, L.; et al. Antibiotic resistance and molecular characterization of the hydrogen sulfide-negative phenotype among diverse Salmonella serovars in China. BMC Infect. Dis 2018, 18, 292. [Google Scholar] [CrossRef] [Green Version]
- Müştak, İ.B.; Müştak, H.K.; Sarıçam, S. Molecular characterisation of hydrogen sulfide negative Salmonella enterica serovar Havana. Antonie Van Leeuwenhoek 2020, 113, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.E.; Alikhan, N.-F.; Dallman, T.J.; Zhou, Z.; Grant, K.; Maiden, M.C.J. Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar Enteritidis outbreak. Int. J. Food Microbiol. 2018, 274, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Alba, P.; Leekitcharoenphon, P.; Carfora, V.; Amoruso, R.; Cordaro, G.; Di Matteo, P.; Ianzano, A.; Iurescia, M.; Diaconu, E.L.; Study Group, E.-E.-A.N.; et al. Molecular epidemiology of Salmonella Infantis in Europe: Insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb. Genom. 2020, 6, e000365. [Google Scholar] [CrossRef] [PubMed]
- Mastrorilli, E.; Pietrucci, D.; Barco, L.; Ammendola, S.; Petrin, S.; Longo, A.; Mantovani, C.; Battistoni, A.; Ricci, A.; Desideri, A.; et al. A comparative genomic analysis provides novel insights into the ecological success of the monophasic Salmonella serovar 4,[5],12:i:. Front. Microbiol. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Hao, X.; Lüthje, F.L.; Qin, Y.; McDevitt, S.F.; Lutay, N.; Hobman, J.L.; Asiani, K.; Soncini, F.C.; German, N.; Zhang, S.; et al. Survival in amoeba—A major selection pressure on the presence of bacterial copper and zinc resistance determinants? Identification of a “copper pathogenicity island.”. Appl. Microbiol. Biotechnol. 2015, 99, 5817–5824. [Google Scholar] [CrossRef]
- Agron, P.G.; Walker, R.L.; Kinde, H.; Sawyer, S.J.; Hayes, D.C.; Wollard, J.; Andersen, G.L. Identification by subtractive hybridization of sequences specific for Salmonella enterica Serovar Enteritidis. Appl Environ Microbiol. 2001, 67, 4984–4991. [Google Scholar] [CrossRef] [Green Version]
- Rasheed, J.K.; Jay, C.; Metchock, B.; Berkowitz, F.; Weigel, L.; Crellin, J.; Steward, C.; Hill, B.; Medeiros, A.A.; Tenover, F.C. Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 1997, 41, 647–653. [Google Scholar] [CrossRef] [Green Version]
- Guerra, B.; Junker, E.; Miko, A.; Helmuth, R.; Mendoza, M.C. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype Typhimurium strains. Microb Drug Resist. 2004, 10, 83–91. [Google Scholar] [CrossRef]
- Kerrn, M.B.; Klemmensen, T.; Frimodt-Moller, N.; Espersen, F. Susceptibility of Danish Escherichia strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulfonamide resistance. J. Antimicrob. Chemother. 2002, 50, 513–516. [Google Scholar] [CrossRef] [Green Version]
- Perreten, V.; Boerlin, P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 2003, 47, 1169–1172. [Google Scholar] [CrossRef] [Green Version]
- Liebert, C.A.; Wireman, J.; Smith, T.; Summers, A.O. Phylogeny of mercury resistance (mer) operons of Gram-negative bacteria isolated from the fecal flora of primates. Appl. Environ. Microbiol. 1997, 63, 1066–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Serotype (no. Isolates)/ST | No. Samples/Farm/Season | Antibiotic Resistance Phenotype/Genotype 1 | Metal Tolerance Genes 2 | MIC Copper Anaerobiosis (mM) | Minimum Growth pH 3 | Minimum Survival pH | MIC Peracetic Acid (mg/L) 3 | MBC Peracetic Acid (mg/L) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| pH Not Adjusted | pH Adjusted to 4.5 | pH Not Adjusted | pH Adjusted to 4.5 | |||||||
| 1,4,[5],12:i:- (n = 6)/ST3478 | 1 sample/farm A/spring | ASSu[T]/ blaTEM, strA-strB, sul2, [tet(B)] | pcoD, silA, arsB, [merA] | 32 | 4.00 | 3.50–4.00 | 60–70 | 40 | 70–90 | 50 |
| Enteritidis (n = 3)/ST11 | 1 sample/farm B/summer | - | - | 4 | 4.00–4.50 | 4.00 | 60–70 | 20–30 | 90 | 40 |
| Serotype (ST - no. Isolates) | Source, Country (Year) | Type of pshA Gene Mutation 1 (Nucleotide, Protein) | Reference (Original Database; Accession no.) 2 |
|---|---|---|---|
| 1,4,[5],12:i:- (3478 - n = 1) | Poultry meat, Portugal (2018) | Nonsense (1669TC > T, Q557X) | This study |
| 1,4,[5],12:i:- (3478 - n = 1) | Human-clinical (small tomatoes, outbreak), Sweden (2019) | Nonsense (1669C > T, Q557X) | [57] (ENA; ERR3577233) |
| Aberdeen (426 - n = 7) | Human-clinical, vegetables, surface water, China (2006–2013) | Nonsense (208C > T, Q70X) | [63] (GenBank; KU143714–KU143732) |
| Choleraesuis (68 - n = 19) | Human-clinical, China (2010–2011) | Frameshift (760delG) | [64] (GenBank; KP184398–184416, 18419–18420) |
| Choleraesuis 4(68 - n = 6) | Human, China (2010, 2012, 2013) | Frameshift (760delG) | [65] (GenBank; KY211936 to KY211941) |
| Havana (1621 - n = 3) | Broiler chickens, Turkey (2016, 2018) | Nonsense (1914C > A, Y638X) | [66] (GenBank; MK548410 to MK548412) |
| Infantis (32 - n = 1) | Poultry meat, Japan (2010) | Nonsense (358G > A, Q120X) | [58] (DDBJ; DRA000592) |
| Paratyphi A (85, 129 - n = 6) | Human, China (2010–2013) | Frameshift (1087delA) | [65] (GenBank; KY211950 to KY211955) |
| Senftenberg (185 - n = 1, 210 - n = 1, 217 - n = 1, 1751 - n = 14) | Seafood product and human-clinical, China (2005–2011) | Nonsense (1621C > T, Q541X) | [60] (GenBank; KF977150 to KF977170) |
| Typhimurium (328 - n = 3) | Poultry meat, Japan (2010) | Nonsense (1440C > A, C480X) | [58] (DDBJ; DRA000592) |
| Typhimurium (1544 - n = 1) | Human, China (2010) | Frameshift (1087delA) | [65] (GenBank; KY211942) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mourão, J.; Rebelo, A.; Ribeiro, S.; Peixe, L.; Novais, C.; Antunes, P. Atypical Non-H2S-Producing Monophasic Salmonella Typhimurium ST3478 Strains from Chicken Meat at Processing Stage Are Adapted to Diverse Stresses. Pathogens 2020, 9, 701. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens9090701
Mourão J, Rebelo A, Ribeiro S, Peixe L, Novais C, Antunes P. Atypical Non-H2S-Producing Monophasic Salmonella Typhimurium ST3478 Strains from Chicken Meat at Processing Stage Are Adapted to Diverse Stresses. Pathogens. 2020; 9(9):701. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens9090701
Chicago/Turabian StyleMourão, Joana, Andreia Rebelo, Sofia Ribeiro, Luísa Peixe, Carla Novais, and Patrícia Antunes. 2020. "Atypical Non-H2S-Producing Monophasic Salmonella Typhimurium ST3478 Strains from Chicken Meat at Processing Stage Are Adapted to Diverse Stresses" Pathogens 9, no. 9: 701. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens9090701