Hydrocarbon-Degrading Bacteria Found Tightly Associated with the 50–70 μm Cell-Size Population of Eukaryotic Phytoplankton in Surface Waters of a Northeast Atlantic Region
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Field Sampling
2.2. Barcoded-Amplicon Illumina MiSeq Sequencing and Bioinformatic Analysis
2.3. FISH-Flow Procedures
2.3.1. Preparation of Pure Cell Culture Controls for FISH-Flow
2.3.2. FISH Protocol
2.3.3. Flow Cytometry
2.3.4. Data Processing and Statistical Analysis
3. Results and Discussion
3.1. MiSeq 16S rRNA Sequencing Analysis of the Bacterial Community Associated with the Eukaryotic Phytoplankton Population in the FSC
3.2. FISH-Flow Optimisation and Methodological Considerations
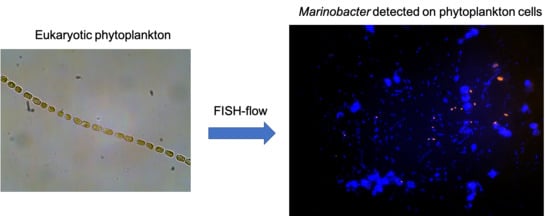
3.3. FISH-Flow Identification of Marinobacter Associated with the Eukaryotic Phytoplankton Population
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Field, C.B.; Behrenfield, M.J.; Randerson, J.T.; Falkowski, P. Primary production of the biosphere: Integrating terrestrial oceanic components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducklow, H.W.; Steinberg, D.K.; Buesseler, K.O. Upper ocean carbon export and the biological pump. Oceanography 2001, 14, 50–58. [Google Scholar] [CrossRef]
- Amin, S.A.; Hmelo, L.R.; van Tol, H.M.; Durham, B.P.; Carlson, L.T.; Heal, K.R.; Morales, R.L.; Berthiaume, C.T.; Parker, M.S.; Djunaedi, B.; et al. Interactions and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 2015, 522, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Parker, M.S.; Ambrust, E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; González, J.M. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 2014, 12, 686–698. [Google Scholar] [CrossRef]
- Amin, S.A.; Green, D.H.; Hart, M.C.; Küpper, F.C.; Sunda, W.G.; Carrano, C.J. Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc. Natl. Acad. Sci. USA 2009, 106, 17071–17076. [Google Scholar] [CrossRef] [Green Version]
- Bell, W.; Mitchell, R. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 1972, 143, 265–277. [Google Scholar] [CrossRef]
- Myklestad, S.M. Release of extracellular products by phytoplankton with special emphasis on polysaccharides. Sci. Total Environ. 1995, 165, 155–164. [Google Scholar] [CrossRef]
- McGenity, T.J.; Folwell, B.D.; McKew, B.A.; Sanni, G.O. Marine crude-oil biodegradation: A central role for interspecies interactions. Aquat. Biosyst. 2012, 8, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazamia, E.; Czesnick, H.; Van Nguyen, T.T.; Croft, M.T.; Sherwood, E.; Sasso, S.; Hodson, S.J.; Warren, M.J.; Smith, A.G. Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ. Microbiol. 2012, 14, 1466–1476. [Google Scholar] [CrossRef]
- Green, D.H.; Bowman, J.P.; Smith, E.A.; Gutierrez, T.; Bolch, C.J.S. Marinobacter algicola sp. nov., isolated from laboratory cultures of paralytic shellfish toxin-producing dinoflagellates. Int. J. Syst. Evol. Microbiol. 2006, 56, 523–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, T.; Nichols, P.D.; Whitman, W.B.; Aitken, M.D. Porticoccus hydrocarbonoclasticus sp. nov., an aromatic hydrocarbon-degrading bacterium identified in laboratory cultures of marine phytoplankton. Appl. Environ. Microbiol. 2012, 78, 628–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, T.; Green, D.H.; Whitman, W.B.; Nichols, P.D.; Semple, K.T.; Aitken, M.D. Algiphilus aromaticivorans gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium isolated from a culture of the marine dinoflagellate Lingulodinium polyedrum, and proposal of Algiphilaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2012, 62, 2743–2749. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, T.; Green, D.H.; Nichols, P.D.; Whitman, W.B.; Semple, K.T.; Aitken, M.D. Polycyclovorans algicola gen. nov., sp. nov., an aromatic-hydrocarbon-degrading marine bacterium found associated with laboratory cultures of marine phytoplankton. Appl. Environ. Microbiol. 2013, 79, 205–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, T.; Rhodes, G.; Mishamandani, S.; Berry, D.; Whitman, W.B.; Nichols, P.D.; Semple, K.T.; Aitken, M.D. Polycyclic aromatic hydrocarbon degradation of phytoplankton-associated Arenibacter spp. and description of Arenibacter algicola sp. nov., an aromatic hydrocarbon-degrading bacterium. Appl. Environ. Microbiol. 2014, 80, 618–628. [Google Scholar] [CrossRef] [Green Version]
- Andelman, J.B.; Suess, M.J. Polynuclear aromatic hydrocarbons in the water environment. Bull. World Health Organ. 1970, 43, 479–508. [Google Scholar]
- Lea-Smith, D.J.; Biller, S.J.; Davey, M.P.; Cotton, C.A.R.; Perez Sepulveda, B.M.; Turchyn, A.V.; Scanlan, D.J.; Smith, A.G.; Chisholm, S.W.; Howe, C.J. Contribution of cyanobacterial alkane production to the ocean hydrocarbon cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 13591–13596. [Google Scholar] [CrossRef] [Green Version]
- Mallet, L.; Sardou, J. Examination of the presence of the polybenzic hydrocarbon benzo-3,4-pyrene in the plank-tonic environment of the Bay of Villefranche. Symposium, committee on international scientific exploration of the mediterranean sea, Monaco. CR Acad. Sci. 1964, 258, 5264–5267. [Google Scholar]
- Exton, D.A.; Steinke, M.; Suggett, D.J.; McGenity, T.J. Spatial and temporal variability of biogenic isoprene emissions from a temperate estuary. Glob. Biogeochem. Cycl. 2012, 26, GB2012. [Google Scholar] [CrossRef] [Green Version]
- Gunnison, D.; Alexander, M. Basis for the resistance of several algae to microbial decomposition. Appl. Microbiol. 1975, 29, 729–738. [Google Scholar] [CrossRef]
- Marlowe, I.T.; Green, J.C.; Neal, A.C.; Brassell, S.C.; Eglinton, G.; Course, P.A. Long chain (n-C37-C39) alkenones in the Prymnesiophyceae. Distribution of alkenones and other lipids and their taxonomic significance. Br. Phycol. J. 1984, 19, 203–216. [Google Scholar] [CrossRef] [Green Version]
- Shaw, S.L.; Gantt, B.; Meskhidze, N. Production and emissions of marine isoprene and monoterpenes: A review. Adv. Meteorol. 2010, 2010, 408696. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Köster, J. The direct role of aerobic heterotrophic bacteria associated with cyanobacteria in the degradation of oil compounds. Int. Biodeterior. Biodegrad. 2005, 55, 29–37. [Google Scholar] [CrossRef]
- Warshawsky, D.; LaDow, K.; Schneider, J. Enhanced degradation of benzo[a]pyrene by Mycobacterium sp. in conjunction with green alga. Chemosphere 2007, 69, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Steen, H.B. Flow cytometer for measurement of the light scattering of viral and other submicroscopic particles. Cytom. A 2004, 57, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, U.; Lenke, J. Improved enumeration of lactic acid bacteria in mesophilic dairy starter cultures by using multiplex quantitative real-time pcr and flow cytometry-fluorescence in situ hybridization. Appl. Environ. Microbiol. 2006, 72, 4163–4171. [Google Scholar] [CrossRef] [Green Version]
- Nettmann, E.; Fröhling, A.; Heeg, K.; Klocke, M.; Schlüter, O.; Mumme, J. Development of a flow-fluorescence in situ hybridization protocol for the analysis of microbial communities in anaerobic fermentation liquor. BMC Microbiol. 2013, 13, 278. [Google Scholar] [CrossRef] [Green Version]
- Wallner, G.; Amann, R.; Beisker, W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 1993, 14, 136–143. [Google Scholar] [CrossRef]
- Li, M.; Yang, Y.; He, Y.; Mathieu, J.; Yu, C.; Li, Q.; Alvarez, P.J.J. Detection and cell sorting of Pseudonocardia species by fluorescence in situ hybridization and flow cytometry using 16S rRNA-targeted oligonucleotide probes. Appl. Microbiol. Biotechnol. 2018, 102, 3375–3386. [Google Scholar] [CrossRef]
- Arrigucci, R.; Bushkin, Y.; Radford, F.; Lakehal, K.; Vir, P.; Pine, R.; Martin, D.; Sugarman, J.; Zhao, Y.; Yap, G.S.; et al. FISH-Flow, a protocol for the concurrent detection of mRNA and protein in single cells using fluorescence in situ hybridization and flow cytometry. Nat. Prot. 2017, 12, 1245–1260. [Google Scholar] [CrossRef] [PubMed]
- Biegala, I.C.; Not, F.; Vaulot, D.; Simon, N. Quantitative assessment of picoeukaryotes in the natural environment by using taxon-specific oligonucleotide probes in association with tyramide signal amplification-fluorescence in situ hybridization and flow cytometry. Appl. Environ. Microbiol. 2003, 69, 5519–5529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekar, R.; Fuchs, B.M.; Amann, R.; Pernthaler, J. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 2004, 70, 6210–6219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manti, A.; Boi, P.; Amalfitano, S.; Puddu, A.; Papa, S. Experimental improvements in combining CARD-FISH and flow cytometry for bacterial cell quantification. J. Microbiol. Methods 2011, 87, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Head, I.M.; Jones, D.M.; Röling, W.F.M. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 2006, 4, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 2007, 18, 257–266. [Google Scholar] [CrossRef]
- Suja, L.D.; Summers, S.; Gutierrez, T. Role of EPS, dispersant and nutrients on the microbial response and MOS formation in the subarctic northeast Atlantic. Front. Microbiol. 2017, 8, 676. [Google Scholar] [CrossRef] [Green Version]
- McKay, L.J.; Gutierrez, T.; Teske, A.P. Development of a group-specific 16S rRNA-targeted probe set for the identification of Marinobacter by fluorescence in situ hybridization. Deep Sea Res. Part II Top. Stud. Oceanogr. 2016, 129, 360–367. [Google Scholar] [CrossRef] [Green Version]
- Berx, B.; Hansen, B.; Østerhus, S.; Larsen, K.M.; Sherwin, T.; Jochumsen, K. Combining in situ measurements and altimetry to estimate volume, heat and salt transport variability through the Faroe–Shetland Channel. Ocean Sci. 2013, 9, 639–654. [Google Scholar] [CrossRef] [Green Version]
- Tillett, D.; Neilan, B. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 2000, 36, 251–258. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Glo, F.O.; Yarza, P. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Ben Mahfoudh, K.; Wagner, M.; Loy, A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol. 2011, 78, 612. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackburn, S.I.; Hallegraeff, G.M.; Bolch, C.J. Vegetative reproduction and sexual life cycle of the toxic dinoflagellate Gymnodinium catenatum from Tasmania. Aust. J. Phycol. 1989, 25, 577–590. [Google Scholar] [CrossRef]
- Porter, K.G.; Feig, Y.S. The use of DAPI for identifyinh and counting aquatic microflora. Limnol. Oceanogr. 1980, 25, 943–948. [Google Scholar] [CrossRef]
- Chernikova, T.N.; Bargiela, R.; Toshchakov, S.V.; Shivaraman, V.; Lunev, E.A.; Yakimov, M.M.; Thomas, D.N.; Golyshin, P.N. Hydrocarbon-degrading bacteria Alcanivorax and Marinobacter associated with microalgae Pavlova lutheri and Nannochloropsis oculata. Front. Microbiol. 2020, 11, 572931. [Google Scholar] [CrossRef]
- Fei, C.; Ochsenkühn, M.A.; Shibi, A.A.; Isaac, A.; Wang, C.; Amin, S.A. Quorum sensing regulates ‘swim-or-stick’ lifestyle in the phycosphere. Environ. Microbiol. 2020, 22, 4761–4778. [Google Scholar] [CrossRef]
- Needham, D.M.; Fuhrman, J.A. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat. Microbiol. 2016, 1, 16005. [Google Scholar] [CrossRef]
- Methé, B.A.; Nelson, K.E.; Deming, J.W.; Momen, B.; Melamud, E.; Zhang, X.; Moult, J.; Madupu, R.; Nelson, W.C.; Dodson, R.J.; et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. USA 2005, 102, 10913–10918. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Nigro, L.M.; Gutierrez, T.; D’Ambrosio, L.; Joye, S.B.; Highsmith, R.; Teske, A.P. Pulsed blooms and persistent oil-degrading bacterial populations in the water column during and after the Deepwater Horizon blowout. Deep Sea Res. Part II Top. Stud. Oceanogr. 2016, 129, 282–291. [Google Scholar] [CrossRef] [Green Version]
- Kleindienst, S.; Seidel, M.; Ziervogel, K.; Grim, S.; Loftis, K.; Harrison, S.; Malkin, S.Y.; Perkins, M.J.; Field, J.; Sogin, M.L. Chemical dispersants can suppress the activity of natural oil-degrading microorganisms. Proc. Natl. Acad. Sci. USA 2015, 112, 14900–14905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suja, L.D.; Chen, X.; Summers, S.; Paterson, D.; Gutierrez, T. Chemical dispersant enhances microbial exopolymer (EPS) production and formation of marine oil/dispersant snow in surface waters of the subarctic northeast Atlantic. Front. Microbiol. 2019, 10, 553. [Google Scholar] [CrossRef] [PubMed]
- Bælum, J.; Borglin, S.; Chakraborty, R.; Fortney, J.L.; Lamendella, R.; Mason, O.U.; Auer, M.; Zemla, M.; Bill, M.; Conrad, M.E.; et al. Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon spill. Environ. Microbiol. 2012, 14, 2405–2416. [Google Scholar] [CrossRef] [PubMed]
- Thompson, H.; Angelova, A.; Bowler, B.; Jones, M.; Gutierrez, T. Enhanced crude oil biodegradative potential of natural phytoplankton-associated hydrocarbonoclastic bacteria. Environ. Microbiol. 2017, 19, 2843–2861. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, H.; Li, R.; Mao, X. Thalassospira xianhensis sp. nov., a polycyclic aromatic hydrocarbon-degrading marine bacterium. Int. J. Syst. Evol. Microbiol. 2010, 60, 1125–1129. [Google Scholar] [CrossRef]
- González, J.M.; Simó, R.; Massana, R.; Covert, J.S.; Casamayor, E.O.; Pedrós-Alió, C.; Moran, M.A. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 2000, 66, 4237–4246. [Google Scholar] [CrossRef] [Green Version]
- Moran, M.A.; Belas, R.; Schell, M.A.; Gonzalez, J.M.; Sun, F.; Sun, S.; Binder, B.J.; Edmonds, J.; Ye, W.; Orcutt, B.; et al. Ecological genomics of marine roseobacters. Appl. Environ. Microbiol. 2007, 73, 4559–4569. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, T. Aerobic hydrocarbon-degrading Gammaproteobacteria–Porticoccus. In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes; Section 6.10; Handbook of Hydrocarbon and Lipid Microbiology; McGenity, T.J., Prince, R., Eds.; Springer: Berlin, Germany, 2018; Volume 6. [Google Scholar]
- Janvier, M.; Grimont, P.A.D. The genus Methylophaga, a new line of descent within phylogenetic branch γ of Proteobacteria. Res. Microbiol. 1995, 146, 543–550. [Google Scholar] [CrossRef]
- Mishamandani, S.; Gutierrez, T.; Aitken, M.D. DNA-based stable isotope probing coupled with cultivation methods implicates Methylophaga in hydrocarbon degradation. Front. Microbiol. 2014, 8, 10. [Google Scholar] [CrossRef]
- Scanlan, D.J.; Ostrowski, M.; Mazard, S.; Dufresne, A.; Garczarek, L.; Hess, W.R.; Post, A.F.; Hagemann, M.; Paulsen, I.; Partensky, F. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 2009, 73, 249–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, T. Occurrence and roles of the obligate hydrocarbonoclastic bacteria in the ocean when there is no obvious hydrocarbon contamination. In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes; Section 6.34; Handbook of Hydrocarbon and Lipid Microbiology; McGenity, T.J., Prince, R., Eds.; Springer: Berlin, Germany, 2018; Volume 6. [Google Scholar]
- Joye, S.; Kostka, J. Microbial Genomics of the Global Ocean System. Earth and Space Science Open Archive. 2020. Available online: https://www.asmscience.org/content/colloquia.58 (accessed on 1 October 2020).
- Lebaron, P.; Catala, P.; Fajon, C.; Joux, F.; Baudart, J.; Bernard, L. A new sensitive, whole-cell hybridization technique for detection of bacteria involving a biotinylated oligonucleotide probe targeting rRNA and tyramide signal amplification. Appl. Environ. Microbiol. 1997, 63, 3274–3278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schönhuber, W.; Fuchs, B.; Juretschko, S.; Amann, R. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 1997, 63, 3268–3273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singer, E.; Webb, E.A.; Nelson, W.C.; Heidelberg, J.F.; Ivanova, N.; Pati, A.; Edwards, K.J. Genomic potential of Marinobacter aquaeolei, a biogeochemical “opportunitroph”. Appl. Environ. Microbiol. 2011, 77, 2763–2771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handley, K.M.; Lloyd, J.R. Biogeochemical implications of the ubiquitous colonization of marine habitats and redox gradients by Marinobacter species. Front. Microbiol. 2013, 4, 136. [Google Scholar] [CrossRef] [Green Version]
- Duran, R. Marinobacter. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Heidelberg, Germany, 2010; pp. 1725–1736. [Google Scholar]
- Mounier, J.; Camus, A.; Mitteau, I.; Vaysse, P.J.; Goulas, P.; Grimaud, R.; Sivadon, P. The marine bacterium Marinobacter hydrocarbonoclasticus SP17 degrades a wide range of lipids and hydrocarbons through the formation of oleolytic biofilms with distinct gene expression profiles. FEMS Microbiol. Ecol. 2014, 90, 816–831. [Google Scholar] [CrossRef] [Green Version]
- Polovina, J.J.; Howell, E.A.; Abecassis, M. Ocean’s least productive waters are expanding. Geophys. Res. Lett. 2008, 35, L03618. [Google Scholar] [CrossRef] [Green Version]
- Signorini, S.R.; Franz, B.A.; McClain, C.R. Chlorophyll variability in the oligotrophic gyres: Mechanisms, seasonality and trends. Front. Mar. Sci. 2015, 2. [Google Scholar] [CrossRef]
- Sebastian, M.; Estrany, M.; Ruiz-Gonzalez, C.; Forn, I.; Sala, M.M.; Gasol, J.M.; Marrase, C. High growth potential of long-term starved deep ocean opportunistic heterotrophic bacteria. Front. Microbiol. 2019, 10, 760. [Google Scholar] [CrossRef]
- Vaysse, P.J.; Sivadon, P.; Goulas, P.; Grimaud, R. Cells dispersed from Marinobacter hydrocarbonoclasticus SP17 biofilm exhibit a specific protein profile associated with a higher ability to reinitiate biofilm development at the hexadecane–water interface. Environ. Microbiol. 2011, 13, 737–746. [Google Scholar] [CrossRef]
- Arnosti, C.; Ziervogel, K.; Yang, T.; Teske, A. Oil-derived marine aggregates—Hot spots of polysaccharide degradation by specialized bacterial communities. Deep Sea Res. Part II Top. Stud. Oceanogr. 2016, 129, 179–186. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, H.F.; Summers, S.; Yuecel, R.; Gutierrez, T. Hydrocarbon-Degrading Bacteria Found Tightly Associated with the 50–70 μm Cell-Size Population of Eukaryotic Phytoplankton in Surface Waters of a Northeast Atlantic Region. Microorganisms 2020, 8, 1955. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8121955
Thompson HF, Summers S, Yuecel R, Gutierrez T. Hydrocarbon-Degrading Bacteria Found Tightly Associated with the 50–70 μm Cell-Size Population of Eukaryotic Phytoplankton in Surface Waters of a Northeast Atlantic Region. Microorganisms. 2020; 8(12):1955. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8121955
Chicago/Turabian StyleThompson, Haydn Frank, Stephen Summers, Raif Yuecel, and Tony Gutierrez. 2020. "Hydrocarbon-Degrading Bacteria Found Tightly Associated with the 50–70 μm Cell-Size Population of Eukaryotic Phytoplankton in Surface Waters of a Northeast Atlantic Region" Microorganisms 8, no. 12: 1955. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms8121955