Microbial Diversity in Subarctic Biocrusts from West Iceland following an Elevation Gradient
Abstract
:1. Introduction
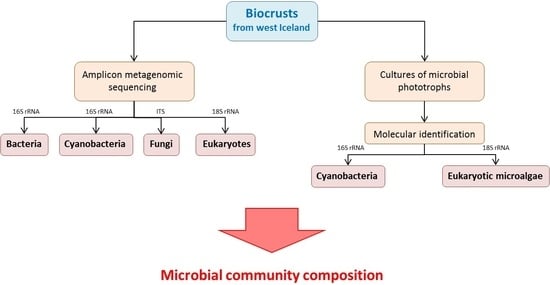
2. Materials and Methods
2.1. Site Description
2.2. Sampling and Soil Characterization
2.3. Cultivation and Molecular Analyses of Microbial Photoautotrophs
2.4. Environmental DNA Extraction and Sequencing
2.5. Bioinformatic and Statistical Analyses
3. Results
3.1. Overall Diversity of Microorganisms in the Icelandic Biocrusts
3.1.1. Bacteria
3.1.2. Cyanobacteria
3.1.3. Fungi
3.1.4. Eukaryotes
3.2. Alpha and Beta Diversities
4. Discussion
4.1. Overall Diversity of Microbiota in Icelandic Biocrusts
4.2. Shift of Microbial Community Composition in Biocrust Depending on Environmental Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Belnap, J.; Lange, O.L. Biological Soil Crusts: Structure, Function, and Management; Springer: Berlin, Germany, 2003; ISBN 9783540437574. [Google Scholar]
- Stewart, K.J.; Lamb, E.G.; Coxson, D.S.; Siciliano, S.D. Bryophyte-cyanobacterial associations as a key factor in N2-fixation across the Canadian Arctic. Plant Soil 2011, 344, 335–346. [Google Scholar] [CrossRef]
- Büdel, B.; Dulić, T.; Darienko, T.; Rybalka, N.; Friedl, T. Cyanobacteria and Algae of Biological Soil Crusts. In Biological Soil Crusts: An Organizing Principle in Drylands. Ecological Studies (Analysis and Synthesis); Weber, B., Büdel, B., Belnap, J., Eds.; Springer: Cham, Switzerland, 2016; Volume 226, pp. 55–80. [Google Scholar]
- Rippin, M.; Borchhardt, N.; Williams, L.; Colesie, C.; Jung, P.; Büdel, B.; Karsten, U.; Becker, B. Genus richness of microalgae and Cyanobacteria in biological soil crusts from Svalbard and Livingston Island: Morphological versus molecular approaches. Polar Biol. 2018, 41, 909–923. [Google Scholar] [CrossRef]
- Steven, B.; Lionard, M.; Kuske, C.R.; Vincent, W.F. High Bacterial Diversity of Biological Soil Crusts in Water Tracks over Permafrost in the High Arctic Polar Desert. PLoS ONE 2013, 8, e71489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, E.; DeLong, E.F.; Lory, S.; Stackebrandt, E.; Thompson, F. (Eds.) The Prokaryotes: Prokaryotic Communities and Ecophysiology; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–528. [Google Scholar] [CrossRef]
- Makhalanyane, T.P.; Valverde, A.; Velázquez, D.; Gunnigle, E.; Van Goethem, M.W.; Quesada, A.; Cowan, D.A. Ecology and biogeochemistry of cyanobacteria in soils, permafrost, aquatic and cryptic polar habitats. Biodivers. Conserv. 2015, 24, 819–840. [Google Scholar] [CrossRef]
- Pichrtová, M.; Kulichová, J.; Holzinger, A. Nitrogen limitation and slow drying induce desiccation tolerance in conjugating green algae (Zygnematophyceae, Streptophyta) from polar habitats. PLoS ONE 2014, 9, e113137. [Google Scholar] [CrossRef]
- Weber, B.; Büdel, B.; Belnap, J. Biological Soil Crusts: An Organizing Principle in Drylands; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Arnalds, Ó. Icelandic soils. Dev. Quat. Sci. 2005, 5, 309–318. [Google Scholar] [CrossRef]
- Opfergelt, S.; Williams, H.M.; Cornelis, J.T.; Guicharnaud, R.A.; Georg, R.B.; Siebert, C.; Gislason, S.R.; Halliday, A.N.; Burton, K.W. Iron and silicon isotope behaviour accompanying weathering in Icelandic soils, and the implications for iron export from peatlands. Geochim. Cosmochim. Acta 2017, 217, 273–291. [Google Scholar] [CrossRef]
- Pushkareva, E.; Baumann, K.; Van, A.T.; Mikhailyuk, T.; Baum, C.; Hrynkiewicz, K.; Demchenko, E.; Thiem, D.; Köpcke, T.; Karsten, U.; et al. Diversity of microbial phototrophs and heterotrophs in Icelandic biocrusts and their role in phosphorus-rich Andosols. Geoderma 2021, 386. [Google Scholar] [CrossRef]
- Byloos, B.; Monsieurs, P.; Mysara, M.; Leys, N.; Boon, N.; Van Houdt, R. Characterization of the bacterial communities on recent Icelandic volcanic deposits of different ages 04 Earth Sciences 0403 Geology. BMC Microbiol. 2018, 18, 1–11. [Google Scholar] [CrossRef]
- Marteinsson, V.; Klonowski, A.; Reynisson, E.; Vannier, P.; Sigurdsson, B.D.; Ólafsson, M. Microbial colonization in diverse surface soil types in Surtsey and diversity analysis of its subsurface microbiota. Biogeosciences 2015, 12, 1191–1203. [Google Scholar] [CrossRef] [Green Version]
- Greipsson, S.; El-Mayas, H.; Vestberg, M.; Walker, C. Arbuscular mycorrhizal fungi in sandy soils in Iceland. Arct. Antarct. Alp. Res. 2002, 34, 419–427. [Google Scholar] [CrossRef]
- Greipsson, S.; El-Mayas, H. Arbuscular mycorrhizae of Leymus arenarius on coastal sands and reclamation sites in Iceland and response to inoculation. Restor. Ecol. 2000, 8, 144–150. [Google Scholar] [CrossRef]
- Eyjolfsdottir, G.G. Soil fungi isolated from Icelandic farmland. Acta Bot. Isl. 1995, 12, 53–62. [Google Scholar]
- Henriksson, L.E.; Henriksson, E. Occurrence of fungi on the volcanic island of Surtsey, Iceland. Acta Bot. Isl. 1974, 3, 82–88. [Google Scholar]
- Broady, P.A. The terrestrial algae of Glerardalur Akureyri, Iceland. Acta Bot. Islandica 1978, 5, 3–60. [Google Scholar]
- Broady, P. Green and yellow-green terrestrial algae from Surtsey (Iceland) in 1978. Surtsey Res. Prog. Rep. 1982, 9, 13–32. [Google Scholar]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high- throughput community sequencing data. Nat. Publ. Gr. 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi-recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. mSystems 2017, 2, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J. Multivariate analysis of ecological communities in R:vegan tutorial. R Doc. 2013, 1–43. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rippin, M.; Lange, S.; Sausen, N.; Becker, B. Biodiversity of biological soil crusts from the Polar Regions revealed by metabarcoding. FEMS Microbiol. Ecol. 2018, 94, 36. [Google Scholar] [CrossRef] [PubMed]
- McCann, C.M.; Wade, M.; Gray, N.D.; Roberts, J.A.; Hubert, C.R.J.; Graham, D.W. Microbial communities in a High Arctic polar desert landscape. Front. Microbiol. 2016, 7, Art 419. [Google Scholar] [CrossRef]
- Malard, L.A.; Pearce, D.A. Minireview Microbial diversity and biogeography in Arctic soils. Environ. Microbiol. Rep. 2018, 10, 611–625. [Google Scholar] [CrossRef]
- Gundlapally, S.R.; Garcia-Pichel, F. The community and phylogenetic diversity of biological soil crusts in the Colorado Plateau studied by molecular fingerprinting and intensive cultivation. Microb. Ecol. 2006, 52, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Mackelprang, R.; Waldrop, M.P.; DeAngelis, K.M.; David, M.M.; Chavarria, K.L.; Blazewicz, S.J.; Rubin, E.M.; Jansson, J.K. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 2011, 480, 368–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Bell, T.H.; Callender, K.L.; Whyte, L.G.; Greer, C.W. Microbial Competition in Polar Soils: A Review of an Understudied but Potentially Important Control on Productivity. Biology 2013, 2, 533–554. [Google Scholar] [CrossRef]
- Arnalds, O.; Hallmark, C.T.; Wilding, L.P. Andisols from four different regions of Iceland. Soil Sci. Soc. Am. J. 1995, 59, 161–169. [Google Scholar] [CrossRef]
- Pushkareva, E.; Pessi, I.S.; Wilmotte, A.; Elster, J. Cyanobacterial community composition in Arctic soil crusts at different stages of development. FEMS Microbiol. Ecol. 2015, 91, fiv143. [Google Scholar] [CrossRef] [PubMed]
- Pushkareva, E.; Wilmotte, A.; Láska, K.; Elster, J. Comparison of Microphototrophic Communities Living in Different Soil Environments in the High Arctic. Front. Ecol. Evol. 2019, 7, 393. [Google Scholar] [CrossRef] [Green Version]
- Raabová, L.; Kovacik, L.; Elster, J.; Strunecký, O. Review of the genus Phormidesmis (Cyanobacteria) based on environmental, morphological, and molecular data with description of a new genus Leptodesmis. Phytotaxa 2019, 395, 1–16. [Google Scholar] [CrossRef]
- Tashyreva, D.; Elster, J. Production of Dormant Stages and Stress Resistance of Polar Cyanobacteria. In Life on Earth and other Planetary Bodies, Cellular Origin, Life in Extreme Habitats and Astrobiology; Hanslmeier, A., Kempe, S., Seckbach, J., Eds.; Springer Science + Business Media: Dordrecht, The Netherlands, 2012; Volume 24, pp. 367–386. ISBN 978-94-007-4965-8. [Google Scholar]
- Robinson, C.H. Cold adaptation in Arctic and Antarctic fungi. New Phytol. 2001, 151, 341–353. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Tedersoo, L.; Abarenkov, K.; Ryberg, M.; Kristiansson, E.; Hartmann, M.; Schoch, C.L.; Nylander, J.A.A.; Bergsten, J.; Porter, T.M.; et al. Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys 2012, 4, 37–63. [Google Scholar] [CrossRef] [Green Version]
- Marañón-Jiménez, S.; Radujković, D.; Verbruggen, E.; Grau, O.; Cuntz, M.; Peñuelas, J.; Richter, A.; Schrumpf, M.; Rebmann, C. Shifts in the Abundances of Saprotrophic and Ectomycorrhizal Fungi With Altered Leaf Litter Inputs. Front. Plant Sci. 2021, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Borchhardt, N.; Baum, C.; Mikhailyuk, T.; Karsten, U. Biological soil crusts of arctic svalbard-water availability as potential controlling factor for microalgal biodiversity. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Rippin, M.; Borchhardt, N.; Karsten, U.; Becker, B. Cold acclimation improves the desiccation stress resilience of polar strains of Klebsormidium (Streptophyta). Front. Microbiol. 2019, 10, 1730. [Google Scholar] [CrossRef] [Green Version]
- Roshan, S.K.; Dumack, K.; Bonkowski, M.; Leinweber, P.; Karsten, U.; Glaser, K. Taxonomic and functional diversity of heterotrophic protists (Cercozoa and endomyxa) from biological soil crusts. Microorganisms 2021, 9, 205. [Google Scholar] [CrossRef]
- Castillo-Monroy, A.P.; Benítez, Á.; Reyes-Bueno, F.; Donoso, D.A.; Cueva, A. Biocrust structure responds to soil variables along a tropical scrubland elevation gradient. J. Arid Environ. 2016, 124, 31–38. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Tamm, A.; Hassenrück, C.; Al-Rawahi, A.N.; Rodríguez-Caballero, E.; Fiedler, S.; Maier, S.; Weber, B. Habitat-dependent composition of bacterial and fungal communities in biological soil crusts from Oman. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef]
- Elster, J. Ecological Classification of Terrestrial Algal Communities of Polar Environment; Springer: Berlin, Germany, 2002. [Google Scholar]
- Canfora, L.; Bacci, G.; Pinzari, F.; Lo Papa, G.; Dazzi, C.; Benedetti, A. Salinity and bacterial diversity: To what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE 2014, 9, e106662. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.; Chan, Y.; Bugler-Lacap, D.C.; Bhatnagar, A.; Bhatnagar, M.; Pointing, S.B. Microbial Diversity in Soil, Sand Dune and Rock Substrates of the Thar Monsoon Desert, India. Indian J. Microbiol. 2016, 56, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Pentecost, A. Order Volvocales. In The Freshwater Algal Flora of the British Isles. An Identification Guide to Freshwater and Terrestrial Algae; John, D., Whitton, B., Brook, A., Eds.; Cambridge University Press: Cambridge, MA, USA, 2002; pp. 303–327. [Google Scholar]
- Smirnova, G.V.; Oktyabrsky, O.N. Glutathione in bacteria. Biokhimiya 2005, 70, 1459–1473. [Google Scholar] [CrossRef] [PubMed]




| (a) | ||
| Code | Site | Nearest Cultivated Neighbor from NCBI Database (ID%) |
| KRA-1a | KRA | MK211225.1 Microcoleus vaginatus KZ-2-2-5 (94.8%) |
| KRA-4b | MK211225.1 Microcoleus vaginatus KZ-2-2-5 (95.1%) | |
| BOR-4c | BOR | KU219728.1 Phormidesmis arctica HOR 11-2 (99.9%) |
| FIF-1a | FIF | KU219728.1 Phormidesmis arctica HOR 11-2 (99.8%) |
| FIF-1d | MK211225.1 Microcoleus vaginatus KZ-2-2-5 (95.3%) | |
| LSK-4c | LSK | MK211225.1 Microcoleus vaginatus KZ-2-2-5 (97.9%) |
| GIL-2a | GIL | KJ939033.1 Phormidesmis sp. WJT36-NPBG15 (97.9%) |
| GIL-2b | MN158649.1 Wilmottia murrayi ACKU582 (98.3%) | |
| GIL-2c | MK211225.1 Microcoleus vaginatus KZ-2-2-5 (98.1%) | |
| GIL-3b | KU219738.1 Phormidesmis arctica MUM 11-8 (97.5%) | |
| (b) | ||
| Code | Site | Nearest Cultivated Neighbor from NCBI Database (ID%) |
| KRA-2b | KRA | JQ315503.1 Chlamydomonas hedleyi KMMCC 188 (99.9%) |
| KRA-4a | MK541759.1 Gonium pectorale CCAP 32/23 (99.3%) | |
| BOR-1c | BOR | AB936289.1 Chlorococcum lobatum (99.0%) |
| BOR-3b | AY846370.1 Ankistrodesmus fusiformis Itas 8/18 M-7w (99.0%) | |
| BOR-3c | MN267184.1 Xerochlorella olmae UTEX B 2993 (99.3%) | |
| BOR-4a | MN248530.1 Chlorella vulgaris CCAP 211/21B (100%) | |
| FIF-1b | FIF | MN267184.1 Xerochlorella olmae UTEX B2993 (100%) |
| FIF-1c | MK541792.1 Chlorella vulgaris CCAP 211/19 (100%) | |
| FIF-2a | MH703776.1 Chlamydomonas callunae SAG 68.81 (99.8%) | |
| FIF-3a | MK262904.1 Klebsormidium elegans ACSSI 187 (100%) | |
| FIF-3b | MN248530.1 Chlorella vulgaris CCAP 211/21B (99.9%) | |
| FIF-4a | LT560349.1 Elliptochloris subsphaerica SAG 2202 (100%) | |
| LSK-1a | LSK | MN267184.1 Xerochlorella olmae UTEX B 2993 (100%) |
| LSK-1b | KM020100.1 Chlorococcum citriforme SAG 62.80 (100%) | |
| LSK-1c | KF673372.1 Chlorella emersonii SAG 2337 (99.9%) | |
| LSK-4a | MN248530.1 Chlorella vulgaris CCAP 211/21B (100%) | |
| LSK-4b | MT425956.1 Heterochlamydomonas sp. ACSSI 328 (99.9%) | |
| GIL-3a | GIL | KJ756841.1 Stichococcus bacillaris CCAP 379/5 (99.0%) |
| GIL-3d | MG696565.1 Nannochloris sp. ACSSI 144 (100%) | |
| GIL-4a | EU878373.1 Parietochloris alveolaris UTEX 836 (99.3%) | |
| r2 | F | |
|---|---|---|
| Bacteria | 0.45 | 3.07 *** |
| Cyanobacteria | 0.58 | 4.75 *** |
| Fungi | 0.41 | 2.59 *** |
| Eukaryotes | 0.54 | 3.47 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pushkareva, E.; Barrantes, I.; Leinweber, P.; Karsten, U. Microbial Diversity in Subarctic Biocrusts from West Iceland following an Elevation Gradient. Microorganisms 2021, 9, 2195. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9112195
Pushkareva E, Barrantes I, Leinweber P, Karsten U. Microbial Diversity in Subarctic Biocrusts from West Iceland following an Elevation Gradient. Microorganisms. 2021; 9(11):2195. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9112195
Chicago/Turabian StylePushkareva, Ekaterina, Israel Barrantes, Peter Leinweber, and Ulf Karsten. 2021. "Microbial Diversity in Subarctic Biocrusts from West Iceland following an Elevation Gradient" Microorganisms 9, no. 11: 2195. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9112195