Supernatants of Bifidobacterium longum and Lactobacillus plantarum Strains Exhibited Antioxidative Effects on A7R5 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Probiotic Strains, Culture Conditions, and Supernatant Preparation
2.2. Cell Culture
2.3. Intracellular ROS Measurement
2.4. Cell Viability Assays
2.5. Intracellular Enzyme Activity Assays
2.6. Total RNA Extraction and RT-qPCR
2.7. Cell Transcriptome Sequencing and Annotation
2.8. Data Processing and Statistical Analysis
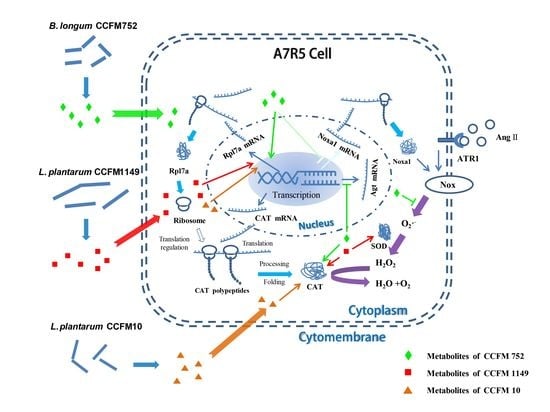
3. Results
3.1. Probiotic Supernatants Inhibited Ang II-Induced ROS Increases in A7R5 Cells
3.2. Probiotic Supernatants Increased the Intracellular Antioxidative Enzyme Activities and Inhibited NADPH Oxidase Activation
3.3. Probiotic Supernatants Did Not Alter Transcriptional Levels of Intracellular Enzymes
3.4. Probiotic Supernatants Altered Transcriptome of A7R5 Cells
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Datla, S.R.; Griendling, K.K. Reactive Oxygen Species, NADPH Oxidases, and Hypertension. Hypertension 2010, 56, 325–330. [Google Scholar] [CrossRef] [Green Version]
- Dikalov, S.I.; Nazarewicz, R.R. Angiotensin II-Induced Production of Mitochondrial Reactive Oxygen Species: Potential Mechanisms and Relevance for Cardiovascular Disease. Antioxid. Redox Signal. 2013, 19, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Byon, C.H.; Heath, J.M.; Chen, Y. Redox signaling in cardiovascular pathophysiology: A focus on hydrogen peroxide and vascular smooth muscle cells. Redox Biol. 2016, 9, 244–253. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.M.; Kim, S.J.; Tatsunami, R.; Yamamura, H.; Fukai, T.; Ushio-Fukai, M. ROS-induced ROS release orchestrated by Nox4, Nox2, and mitochondria in VEGF signaling and angiogenesis. Am. J. Physiol. Cell Physiol. 2017, 312, C749–C764. [Google Scholar] [CrossRef] [Green Version]
- Torrecillas, G.; Boyano-Ad’anez, M.C.; Medina, J.; Parra, T.; Griera, M.; López-Ongil, S.; Arilla, E.; RodrÍguez-Puyol, M.; RodrÍguez-Puyol, D. The role of hydrogen peroxide in the contractile response to angiotensin II. Mol. Pharm. 2001, 59, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Te Riet, L.; van Esch, J.H.; Roks, A.J.; van den Meiracker, A.H.; Jan Danser, A.H. Hypertension: Renin-angiotensin-aldosterone system alterations. Circ. Res. 2015, 116, 960–975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Griendling, K.K.; Dikalova, A.; Owens, G.K.; Taylor, W.R. Vascular hypertrophy in angiotensin II-induced hypertension is mediated by vascular smooth muscle cell-derived H2O2. Hypertension 2005, 46, 732–737. [Google Scholar] [CrossRef] [Green Version]
- Briones, A.M.; Rodríguez-Criado, N.; Hernanz, R.; García-Redondo, A.B.; Rodrigues-Díez, R.R.; Alonso, M.a.J.; Egido, J.S.; Ruiz-Ortega, M.; Salaices, M. Atorvastatin Prevents Angiotensin II–Induced Vascular Remodeling and Oxidative Stress. Hypertension 2009, 54, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Saleh, M.A.; Kirabo, A.; Itani, H.A.; Montaniel, K.R.C.; Xiao, L.; Chen, W.; Mernaugh, R.L.; Cai, H.; Bernstein, K.E.; et al. Immune activation caused by vascular oxidation promotes fibrosis and hypertension. J. Clin. Investig. 2015, 126, 50–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davinelli, S.; Nielsen, M.E.; Scapagnini, G. Astaxanthin in Skin Health, Repair, and Disease A Comprehensive Review. Nutrients 2018, 10, 522. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, F.U.; Sattar, M.A.; Rathore, H.A.; Tan, Y.C.; Akhtar, S.; Jin, O.H.; Pei, Y.P.; Abdullah, N.A.; Johns, E.J. Hydrogen sulphide and tempol treatments improve the blood pressure and renal excretory responses in spontaneously hypertensive rats. Ren Fail. 2014, 36, 598–605. [Google Scholar] [CrossRef]
- Virdis, A.; Gesi, M.; Taddei, S. Impact of apocynin on vascular disease in hypertension. Vasc. Pharm. 2016, 87, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Malekmohammad, K.; Sewell, R.D.E.; Rafieian-Kopaei, M. Antioxidants and Atherosclerosis: Mechanistic Aspects. Biomolecules 2019, 9, 301. [Google Scholar] [CrossRef] [Green Version]
- Miremadi, F.; Ayyash, M.; Sherkat, F.; Stojanovska, L. Cholesterol reduction mechanisms and fatty acid composition of cellular membranes of probiotic Lactobacilli and Bifidobacteria. J. Funct. Foods 2014, 9, 295–305. [Google Scholar] [CrossRef]
- Robles-Vera, I.; Toral, M.; la Visitación, N.; Sánchez, M.; Gómez-Guzmán, M.; Romero, M.; Yang, T.; Izquierdo-Garcia, J.L.; Jiménez, R.; Ruiz-Cabello, J.; et al. Probiotics Prevent Dysbiosis and the Rise in Blood Pressure in Genetic Hypertension: Role of Short-Chain Fatty Acids. Mol. Nutr. Food Res. 2020, 64. [Google Scholar] [CrossRef]
- Han, C.; Ding, Z.; Shi, H.; Qian, W.; Hou, X.; Lin, R. The Role of Probiotics in Lipopolysaccharide-Induced Autophagy in Intestinal Epithelial Cells. Cell Physiol. Biochem. 2016, 38, 2464–2478. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yi, R.; Zhou, X.; Mu, J.; Long, X.; Pan, Y.; Song, J.; Park, K. Preventive effect of Lactobacillus plantarum KSFY02 isolated from naturally fermented yogurt from Xinjiang, China, on d-galactose-induced oxidative aging in mice. J. Dairy Sci. 2019, 102, 5899–5912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Shang, N.; Li, P. In vitro and in vivo antioxidant activity of exopolysaccharide fractions from Bifidobacterium animalis RH. Anaerobe 2011, 17, 226–231. [Google Scholar] [CrossRef]
- Xing, J.; Wang, G.; Zhang, Q.; Liu, X.; Yin, B.; Fang, D.; Zhao, J.; Zhang, H.; Chen, Y.Q.; Chen, W. Determining antioxidant activities of lactobacilli by cellular antioxidant assay in mammal cells. J. Funct. Foods 2015, 19, 554–562. [Google Scholar] [CrossRef]
- Liu, Z.; Dong, L.; Jia, K.; Zhan, H.; Zhang, Z.; Shah, N.P.; Tao, X. Sulfonation of Lactobacillus plantarum WLPL04 exopolysaccharide amplifies its antioxidant activities in vitro and in a Caco2 cell model. J. Dairy Sci. 2019, 102, 5922–5932. [Google Scholar] [CrossRef]
- Achuthan, A.A.; Duary, R.K.; Madathil, A.; Panwar, H.; Kumar, H.; Batish, V.K.; Grover, S. Antioxidative potential of lactobacilli isolated from the gut of Indian people. Mol. Biol. Rep. 2012, 39, 7887–7897. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Madamanchi, N.R.; Vendrov, A.E.; Tchivilev, I.; Rojas, M.; Madamanchi, C.; Brandes, R.P.; Krause, K.H.; Humphries, J.; Smith, A.; et al. Nox Activator 1 A Potential Target for Modulation of Vascular Reactive Oxygen Species in Atherosclerotic Arteries. Circulation. 2010, 121, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Zafari, A.M.; Ushio-Fukai, M.; Akers, M.; Yin, Q.; Shah, A.; Harrison, D.G.; Taylor, W.R.; Griendling, K.K. Role of NADH/NADPH oxidase–derived H2O2 in angiotensin II–induced vascular hypertrophy. Hypertension 1998, 32, 488–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griendling, K.K.; Minieri, C.A.; Ollerenshaw, J.D.; Alexander, R.W. Angiotensin II Stimulates NADH and NADPH Oxidase Activity in Cultured Vascular Smooth Muscle Cells. Circ. Res. 1994, 74, 1141–1148. [Google Scholar] [CrossRef] [Green Version]
- Guzik, T.J.; Touyz, R.M. Oxidative Stress, Inflammation, and Vascular Aging in Hypertension. Hypertension 2017, 70, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Honjo, T.; Otsui, K.; Shiraki, R.; Kawashima, S.; Sawamura, T.; Yokoyama, M.; Inoue, N. Essential Role of NOXA1 in Generation of Reactive Oxygen Species Induced by Oxidized Low-Density Lipoprotein in Human Vascular Endothelial Cells. Endothelium. 2009, 15, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Gerst, J.E. Pimp My Ribosome: Ribosomal Protein Paralogs Specify Translational Control. Trends Genet. 2018, 34, 832–845. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, C.; Zamocky, M.; Sandoval, J.M.; Verrax, J.; Calderon, P.B. Regulation of catalase expression in healthy and cancerous cells. Free Radic. Biol. Med. 2015, 87, 84–97. [Google Scholar] [CrossRef]
- Wang, W.; Xia, M.X.; Chen, J.; Yuan, R.; Deng, F.N.; Shen, F.F. Gene expression characteristics and regulation mechanisms of superoxide dismutase and its physiological roles in plants under stress. Biochemistry 2016, 81, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Babiano, R.; Badis, G.; Saveanu, C.; Namane, A.; Doyen, A.; Díaz-Quintana, A.; Jacquier, A.; Fromont-Racine, M.; de la Cruz, J. Yeast ribosomal protein L7 and its homologue Rlp7 are simultaneously present at distinct sites on pre-60S ribosomal particles. Nucleic Acids Res. 2013, 41, 9461–9470. [Google Scholar] [CrossRef]
- Wu, S.; De Croos, J.N.A.; Storey, K.B. Cold acclimation-induced up-regulation of the ribosomal protein L7 gene in the freeze tolerant wood frog, Rana sylvatica. Gene 2008, 424, 48–55. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Omranian, N.; Ahmadi, F.S.; Nikoloski, Z.; Mueller-Roeber, B. Effect of salt stress on genes encoding translation-associated proteins inArabidopsis thaliana. Plant Signal. Behav. 2014, 7, 1095–1102. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.H.; Mohammadmoradi, S.; Chen, J.Z.; Sawada, H.; Daugherty, A.; Lu, H.S. Renin-Angiotensin System and Cardiovascular Functions. Arter. Thromb. Vasc. Biol. 2018, 38. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Fang, Z.; Zhai, Q.; Cui, S.; Zhao, J.; Zhang, H.; Chen, W.; Lu, W. Supernatants of Bifidobacterium longum and Lactobacillus plantarum Strains Exhibited Antioxidative Effects on A7R5 Cells. Microorganisms 2021, 9, 452. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9020452
Wang Y, Fang Z, Zhai Q, Cui S, Zhao J, Zhang H, Chen W, Lu W. Supernatants of Bifidobacterium longum and Lactobacillus plantarum Strains Exhibited Antioxidative Effects on A7R5 Cells. Microorganisms. 2021; 9(2):452. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9020452
Chicago/Turabian StyleWang, Yusheng, Zhifeng Fang, Qixiao Zhai, Shumao Cui, Jianxin Zhao, Hao Zhang, Wei Chen, and Wenwei Lu. 2021. "Supernatants of Bifidobacterium longum and Lactobacillus plantarum Strains Exhibited Antioxidative Effects on A7R5 Cells" Microorganisms 9, no. 2: 452. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms9020452