1. Introduction
Although part of the normal microbiome in humans,
Candida albicans (
C. albicans) can switch from a harmless commensal fungus to a human pathogen causing infections that range from superficial oral or vaginal candidiasis to life-threatening systemic infections [
1]. Risk factors of
Candida infections include diabetes, pregnancy, neutropenia, and immunocompromization following intravenous catheterization, chemotherapy, transplantation, invasive procedures, prolonged use of broad-spectrum antibiotics, or AIDS [
2,
3].
C. albicans is classified as the major etiological agent of invasive fungal infections [
4]. It is considered a critical public health concern as it imposes economic and medical burdens such as high costs of care, high mortality rate, and extended hospitalization duration [
5].
Typically, treatment of
C. albicans infections is limited to five major classes of antifungal drugs: azoles, polyenes, fluoropyrimidines, allylamines, and echinocandins [
6]. Azole antifungals are usually the most preferred drugs for treating candidiasis as they have limited toxicity, are relatively cheap, and are administered orally [
7]. The azole drug fluconazole is the most commonly used antifungal [
8]. As with other azoles, fluconazole suppresses lanosterol 14-α-demethylase [
9]. This enzyme is encoded by
ERG11 and normally functions in catalyzing lanosterol to C14-demethyl-lanosterol and consequently synthesizing ergosterol which is a main component of the cell membrane [
10]. By inhibiting the demethylation of lanosterol, fluconazole inhibits ergosterol biosynthesis and causes the accumulation of methylated sterols in the cellular membrane, thereby disrupting the cell membrane and arresting fungal growth [
11,
12]. Since fluconazole is fungistatic and not fungicidal, treatment permitted the development of acquired resistance among some
C. albicans strains in the presence of fluconazole [
13]. The prolonged and widespread use of fluconazole, and azoles in general, increases the risk of multidrug resistance that is a critical concern in antifungal therapy [
14].
The onset of resistance to antifungals is clinically important and is controlled by several mechanisms. For instance, point mutations in
ERG11 result in the reduced ability of the enzyme to bind fluconazole [
15]. Another mechanism is through the overexpression of
ERG11 leading to increased ergosterol biosynthesis [
16]. Furthermore, gain-of-function gene mutations in some transcription factors lead to constitutive up regulation of numerous target genes and increased fluconazole resistance. For example, mutations in Mrr1 and Tac1 constitutively overexpress the efflux pumps
MDR1 and
CDR1/
CDR2, respectively, which considerably enhance fluconazole resistance in
C. albicans through decreasing intracellular drug accumulation [
17]. Additional mechanisms include alterations in enzymes involved in the ergosterol biosynthetic pathway, changes in plasma membrane components and cell wall proteins (CWPs) types, and lowered uptake of fluconazole, in addition to other mechanisms yet to be elucidated [
18].
We have previously obtained drug-resistant and sensitive
C. albicans isolates from local Lebanese medical centers. One of these strains was found to be resistant to all azoles tested [
19]. We recently subjected the strain to multiple phenotypic assays to characterize it. The strain exhibited reduced growth, a 26% increase in chitin content, 70% increase in membrane ergosterol content, and a 44% decrease in biofilm formation when compared to the reference
C. albicans strain SC5314. It was also found to be overly adherent, but with a decreased resistance to cell wall-disrupting agents.
ERG11 sequencing indicated frame-shift mutations and a disseminated virulence assay revealed that the strain was slightly attenuated in virulence compared to the reference strains [
20]. Since fluconazole resistance is the result of the up-regulation of efflux pumps that are localized to the cell surface and differential expression of many CWPs, we applied a proteomic approach to our fluconazole-resistant
C. albicans strain to identify CWPs expressed upon exposure to fluconazole. Such CWP might explain the mechanisms behind the above-mentioned phenotypes. This pilot study will be expanded in the future to include fluconazole-sensitive strains and compare their proteomic profiles to fluconazole resistant ones with the aim of detecting differentially expressed proteins involved in resistance acquisition mechanisms.
2. Materials and Methods
Strains used: One fluconazole-resistant
C. albicans strain (MIC > 256 μg/mL) was utilized in this study [
19].
Culture Conditions: RPMI 1640 media (with L-glutamine and no bicarbonate) with MOPS and glucose (AB Biodisks) was prepared according to the manufacturer’s instructions. The strain was grown at 28°C on potato dextrose agar plates (Oxoid) supplemented with histidine and uridine. C. albicans colonies were suspended in 0.154 M sterile saline solution up to a turbidity of 0.5 McFarlans and were spread over RPMI 1640 plates with a sterile cotton swab. A fluconazole E-test strip (AB Biodisk, Solna, Sweden) was then placed at the center of each RPMI 1640 plate and the plates were incubated for 48 h at 28 °C.
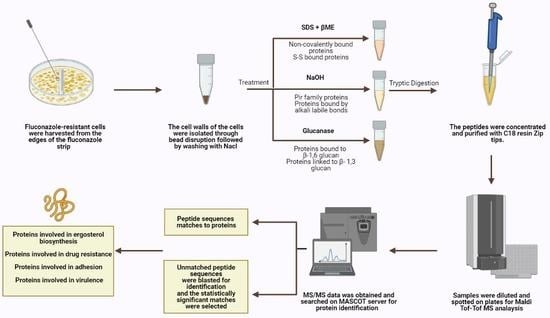
Cell harvesting: Fluconazole-resistant cells were harvested from the edges of the fluconazole strip outwards at high concentrations, ensuring that the harvested cells were properly exposed to the antifungal drug.
Cell wall isolation and protein extraction: Eight independent cell wall extractions were performed as described previously [
21]. A minimum weight of 50 mg of cultured cells was collected and the cells were washed with water at least three times, centrifuged at 4000 rpm for 5 min, and then re-suspended in 5 mL Tris (5 mM, pH = 7.8). They were then divided into 1 mL Eppendorf tubes and centrifuged for 3 min at 12,000 rpm. Pellets were resuspended in 750 μL Tris (5 mM, pH = 7.8). Protease inhibitor cocktail (6 μL, Abcam ab65621) and cold glass beads were added (beads-to-pellets ratio of 1:1). Thirty vortexing cycles on a bead beater were performed for breakage as follows: 30 s on vortex then 30 s on ice. Orange color resulting from the reaction between the acidic cytosol and the protease inhibitor signifies breakage and the efficiency of the breakage was monitored using microscopy. The solution on top of the beads was transferred to new pre-weighed tubes. The beads were washed multiple times using cold NaCl (1 mM) and poured again over the respective pre-weighed tubes to collect as much cell wall material as possible without transferring any beads during the procedure. Samples were centrifuged at 3000 rpm for 5 min. The supernatant containing intracellular proteins was poured off while the pellet was washed in NaCl (40 mL, 1 mM) at least three times. SDS extraction buffer (50 mM Tris, 2% SDS, 100 mM Na-EDTA, 150 mM NaCl, pH 7.8) with β-mercaptoethanol (β-ME) (8 μL per 1 mL SDS extraction buffer) was added (0.5 mL buffer per 100 mg wet weight walls). Tubes were boiled for 10 min then cooled down to room temperature. They were centrifuged for 5 min at 3000 rpm and the supernatant was collected and labeled SDS-extracted protein fraction to be later subjected to tryptic digestion. SDS extraction buffer and β-ME were added as before to resuspend the pellet. The tubes were boiled and cooled down to room temperature three more times then centrifuged for 5 min at 3000 rpm and the resulting pellet was suspended and washed with water to remove excess SDS. The final pellet was frozen in liquid nitrogen and freeze-dried. Lyophilized cell walls were stored at 20 °C until use.
Extraction of alkali labile CWPs: Cell wall pellets were incubated overnight with NaOH (30 mM) at 4 °C and then neutralized with aqueous acetic acid (30 mM) [
22]. They were centrifuged; supernatants were collected and subjected to tryptic digestion.
Extraction of CWPs using Glucanase: Cell wall pellets were treated with 1 mg of B-(1-3)-D-Glucanase from
Helix pomatia (Sigma-Aldrich 67138, specific activity ≥ 0.2 U/mg) in sodium acetate buffer (1 mL, 150 mM, pH = 5) per 10
8 cell equivalents and were incubated overnight at 37 °C [
23]. Cell numbers were estimated using spectrophotometric analysis at 600 nm. Supernatants were collected and subjected to tryptic digestion.
Tryptic digestion: CWP fractions were incubated in a reducing buffer (10 mM DTT, 100 mM NH
4HCO
3) at 55 °C for an hour then spun and pellets were resuspended in an alkylating buffer (65 mM iodoacetamide, 100 mM NH
4HCO
3). They were incubated for 45 min at room temperature in the dark. A quenching solution (55 mM DTT, 100 mM NH
4HCO
3) was added to the samples and kept for five minutes at room temperature followed by 5× washing with ammonium bicarbonate buffer (50 mM). The pellets were re-suspended in a solution containing ammonium bicarbonate (50 mM) and trypsin (1 μg/μL) and left at 37 °C for 16 h. Finally, they were spun, and the supernatants were collected. TFA (0.1%
v/v) was added to prepare samples for subsequent ZipTip clean-up [
24].
ZipTip clean-up: ZipTip C18 clean-up tips (Millipore® Ziptips, Sigma-Aldrich, volume 10 μL, 0.6 μL C18 resin) were wetted in acetonitrile solution. They were equilibrated in HPLC water solution containing 0.1% TFA. Sample binding was achieved by full pressing at least 10 times in the sample tubes. The tips were then washed in HPLC water solution containing 0.1% TFA. The samples were eluted using 10 μL of elution buffer (0.1% TFA (v/v) in HPLC water/acetonitrile (1:1)).
Tandem mass spectrometry: Concentrated and cleaned-up peptides were spotted on a stainless-steel target plate (Opti-TOF TM 384 Well Insert, 128 × 81 mm RevA, Applied Biosystems). They were overlaid with α-cyano-4-hydroxy-cinnamic acid matrix solution (10 mg CHCA matrix in 50% acetonitrile with 0.1% TFA) and air-dried. MALDI-TOF-TOF MS spectra were acquired using the 4800 MALDI-TOF-TOF analyzer, operated by the 4000 Series Explorer software (version 3.7). The instrument was externally calibrated using TOF/TOF Calibration Mixture (Mass Standards Kit for Calibration of AB SCIEX TOF/TOF Instruments). MS reflector positive mode was used for acquisition at a laser intensity of 2500. The mass range was set at 499 to 2500 Da and the focus mass was set at 1500 Da. The reflector positive default was used as a processing method with a minimum signal-to-noise ratio of 5. The resulting mass lists were manually scanned for known contaminant mass peaks (keratin, matrix, and trypsin autolysis) and an exclusion list was created to be applied for all performed MS/MS data acquisitions. For the interpretation method, MS/MS 1 kV positive was utilized as an MS/MS acquisition method specifying a fixed laser intensity of 3500 and a precursor mass of 1570.677 Da. Metastable suppressor and CID were turned on. MS/MS positive default was utilized as an MS/MS processing method in which a signal-to-noise threshold of 5 for monoisotopic peaks was chosen.
Protein identification: MS/MS Ion Search was performed first using the common Repository of Adventitious Proteins, cRAP, database to eliminate possible contaminants whose peaks were not inserted in the exclusion list. Then, a custom
C. albicans CWP database on the MASCOT server was used in order to identify the proteins within the samples as described previously [
24]. The database includes sequences of all curated
C. albicans proteins available in the Swissprot database with gene ontology related to cell wall, plasma membrane, and transmembrane localization tags. As such, the database contains a total of 218 sequences to be searched. In the MASCOT Server’s search parameters page, peptide and fragment tolerance values were set at 2 Da each, “Carbamidomethyl C” and “Oxidation M” were chosen as fixed modification and variable modification, respectively, and up to two missed cleavages were allowed for trypsin. Moreover, a peptide charge of 1+ was allocated and MALDI-TOF-TOF was selected in the instrument type option. In the peptide summary report obtained by MASCOT, individual ions scores greater than 15 indicated identity or extensive homology and proteins were considered successful hits if they had a minimum sequence coverage of 2% or if their peptide sequence was at least 12 amino acids long as these are sufficient for unmistakable protein identification [
25]. Peptide sequences provided by MASCOT but unassigned to any protein were identified via BLAST search in the
Candida Genome database (
www.candidagenome.org (accessed on 10 January 2021) in which the chosen target genome and target sequence dataset were “
Candida albicans SC5314 Assembly 22” and “Proteins–translation of coding sequence (PROTEIN)”, respectively, and no gapped alignments were permitted. The cutoff E-value was set at <0.05 for both MASCOT and BLAST searches. The STRING database (version 11) was used to find a protein–protein interaction prediction map of detected proteins [
26]. The confidence score and false-discovery rate (FDR) stringency were set at 0.400 and 5%, respectively, and the chosen organism was
Candida albicans. The STRING database was also used to highlight proteins with the following biological processes: drug metabolic process (GO: 0017144), cellular response to drug (GO: 0035690), response to fungicide (GO: 0060992), and/or ergosterol biosynthetic pathway (GO: 0006696).
4. Discussion
The cell wall in a fungus is antigenic as it is the first structure that contacts the host. It includes and incorporates proteins involved in adhesion, biofilm formation, virulence and drug resistance [
24]. This study aimed to determine the cell wall proteomic profile of a previously characterized fluconazole-resistant
C. albicans strain exposed to fluconazole in order to identify proteins involved in azole resistance and pathogenicity-related phenotypes. We have previously successfully applied such a technique in identifying proteins differentially expressed in cell wall mutants [
21,
24]. In this context, we are applying it to determine changes in cell wall protein composition in response to drug exposure. The previous characterization of our resistant strain found a 44% reduction in biofilm formation, a 26% increase in chitin content, and a 70% increase in ergosterol content compared to the reference strain [
20]. The strain was also hyper-adherent, and slightly less virulent. Since the cell wall is the initial point of contact between the fungus and its host, its composition is dynamic, and CWP expression varies depending on specific environmental stresses and cues [
27]. We hypothesized that a fluconazole-resistant strain under fluconazole exposure results in the expression of proteins and in changes at the level of the cell wall structure that explain our previously observed phenotypes. CWP extraction following fluconazole exposure coupled with tandem mass spectrometry and database mining revealed such proteins.
In total, 50 proteins associated with the cell surface were identified in this study. Interestingly, we were able to identify some atypical proteins otherwise referred to as moonlighting proteins since they are of cytoplasmic origin yet are also detected at the cell surface with a different yet important role in fungal virulence such as Fba1, Gpm1, Ipp1, and Eno1 [
28]. Gpm1 has been reported to be an important virulence factor in
C. albicans as it is involved in degradation of the extracellular matrix and host immune evasion and was found to be induced by fluconazole [
29,
30]. Ipp1 and Eno1 are also essential proteins. The latter has a role in the growth, morphogenesis, cell division and osmotic protection of the fungus [
31]. Another atypical protein is Tsa1B, a peroxidase that plays a major role in resisting heat shock and oxidative stress [
32]. Additionally, proteins with known functions in the mitochondria (Pam17, Ssc1, Mic60, and Aim36) were detected; they could either be artifacts resulting from contamination during cell wall isolation or atypical proteins with unclear functions in the cell wall. The essential protein, Eft2, is antigenic and was assigned as an atypical protein in a previous study [
33]. It is induced upon exposure to stresses and is the target of sordarin antifungals [
34]. Additional essential proteins were identified such as Ape2, Pan1, and Hsp90. Proteins involved in cytokinesis, budding, glycolysis or other processes required for growth were also detected: Eng1, Bud4, and Mts1. The identified heat shock proteins (Hsp70 and Hsp90) play roles in multiple cellular pathways and consequently confer drug resistance [
35]. These proteins identified in the fluconazole-resistant strain explain its ability to survive in the presence of fluconazole. Yet, the low number of detected essential proteins could explain its growth at a lower rate.
Proteins promoting adhesion were detected in our fluconazole-resistant strain such as Big1, Pga60, and Als1. One of the most important adhesins in
C. albicans, Als1, is crucial for fungal adherence and aggregation and was recently linked to fluconazole resistance [
36]. In addition to their roles in cell wall assembly, the glycosidases Utr2 and Crh11 were found to be involved in the adhesion of
C. albicans to host cells [
37,
38]. As such, these proteins explain the adherent phenotype previously observed in the studied strain [
20]. However, Ywp1, which is an anti-adhesive protein only expressed in the yeast form and thought to function in yeast cell dispersal to colonize new sites, was also detected [
39]. Yeast cells lacking this protein were found to form thicker biofilms than their counterparts [
40]. This could explain the previously observed 44% decreased biofilm ability of our fluconazole-resistant strain. Another protein localized to yeast cells and not to hyphae is Bmh1. Knowing that fluconazole hinders hyphal formation, the observed phenotype of reduced biofilm formation ability is not surprising especially since biofilm formation requires both yeast and hyphal forms [
41]. The studied fluconazole-resistant strain could still form biofilms due to the presence of Csa1and Pga10, which play a crucial role in biofilm formation, development and maintenance [
42].
Plasma membrane fluidity is increased following fluconazole treatment which could affect the integrity of the cell wall and lead to increased chitin content as a compensation; our strain was found to exhibit a disrupted cell wall following SDS treatment and a 26% increase in chitin content [
20]. The identified glycosidases (Crh11 and Utr2) and pH-responsive proteins (Phr1 and Phr2) are induced by fluconazole exposure and they are involved in cell wall cross-linking and cell wall integrity [
38,
43]. Furthermore, the identified proteins involved in endocytosis (Pil1 and End3) could explain the increased fluidity caused by fluconazole exposure [
43]. In addition, Pil1 is involved in echinocandin binding. Its presence here is interesting since our strain is sensitive to the echinocandin caspofungin, suggesting a possible role for Pil1 in azole resistance. The septin Cdc11 is involved in chitin deposition and the chitinase Cht4 functions in chitin remodeling [
44,
45], thus explaining the increased chitin content in our strain.
The fungal cell wall usually incorporates many virulence factors including lipases, secreted aspartyl proteases, and superoxide dismutases that enable the fungus to acquire nutrients from its extracellular environment, invade the host, and escape host defenses [
1]. In our strain, we only detected Lip9 and Sap5. In addition, the transcriptional regulators Stp2 and Stp3 that activate genes encoding lipases were detected and are involved in virulence [
46]. The low number of virulence genes detected and the absence of other key virulence proteins could be the reason for the reduced virulence ability of the strain.
We previously demonstrated that the resistant-fluconazole
C. albicans strain had a frameshift in
ERG11 generating a protein that cannot bind to fluconazole and resulting in resistance. Erg11 overexpression might be another mechanism contributing to fluconazole resistance in our strain, especially since Erg11 was detected with relatively high coverage. In addition to Erg11, Erg6 and Bna4, which play roles in ergosterol biosynthesis, were also detected. This is in agreement with studies showing an increased abundance of Erg6 in fluconazole-resistant
C. albicans strains upon exposure to fluconazole [
47]. Since these proteins function in ergosterol biosynthesis, their presence explains the observed 70% increase in ergosterol content. It has been suggested that the need for iron increases during azole stress, especially given that iron is required by multiple enzymes acting in the ergosterol biosynthetic pathway [
43]. Csa1 and Rbt8 are two identified proteins functioning in heme-iron utilization [
42]. Another mechanism for acquiring fluconazole resistance is through incorporating efflux pumps [
17]. We were able to detect two major efflux pumps (Mdr1 and Cdr1) in our studied fluconazole-resistant strain. Mdr1 is a major facilitator superfamily (MFS) efflux pump while Cdr1 is a pleiotropic ATP-binding cassette (ABC) efflux transporter; they transport azoles out of the cell, thus reducing drug accumulation intracellularly and permitting resistance [
48,
49].
Moreover, we found that some of the identified proteins are involved in interrelated pathways such as the biosynthesis of steroids and antibiotics, glycolysis, and metabolic pathways. We were also able to identify some proteins involved in the drug metabolic process (Eno1, Cht4, Gpm1, Fba1, and Bna4), cellular response to drug (Stp2, Stp3, Eft2, Hsp90, Phr1, Sec4, Erg6, Erg11, Mdr1, and Cdr1), response to fungicide (Cdr1 and Mdr1), and ergosterol biosynthetic pathway (Erg6 and Erg11).
For a comparison between fluconazole-resistant and sensitive
C. albicans strains, we present the preliminary MASCOT results of the same cell wall extraction method applied on one sensitive strain. A total of 20 proteins were identified, four of which were common with the studied fluconazole-resistant strain: Hsp90, Phr1, Sec4, and Pam17. Since both strains are grown in the presence of fluconazole and knowing that Hsp90 and Phr1 are part of the cellular response to the drug, the detection of these two proteins in both strains is not surprising. Some of the proteins solely detected in the sensitive strain were constituents of the mitochondria and endoplasmic reticulum, such as Pam18 and Irc22-1, respectively. Rps1 is a constituent of the ribosome and was shown to elicit a host antibody response during infection [
50]. Additionally, we detected Kex2 in the fluconazole-sensitive strain, which is a subtilisin-like protease that processes the aspartyl proteinase Sap2 and is needed for virulence and hyphal growth of
C. albicans [
51]. Several important mannosyltransferases were detected solely in the fluconazole-sensitive strain, such as Alg1, Rhd1, Mnn1, Mnn12, and Mnn13. The proteins Alg1 and Rhd1 are beta-mannosyltransferases needed for growth and cell wall biosynthesis [
52,
53]. Mnn1, Mnn12, and Mnn13 are alpha-mannosyltransferases involved in hyphal development, biofilm formation, and virulence [
54]. Manipulation of the mannan layer and composition increases cell-to-cell adhesion and provides protection from environmental stresses such as antifungals [
55]. These proteins were present in the fluconazole-sensitive strain and therefore explain its increased virulence, colonization, and biofilm forming ability when compared to the fluconazole-resistant strain. Likewise, their lack of detection in the fluconazole-resistant strain could explain its attenuated virulence and defect in biofilm formation. Another important protein detected exclusively in the sensitive strain is Pga14. This protein is a hydrophilin, induced during cell wall regeneration and it is critical for overcoming the stress of desiccation-rehydration process [
56]. The remaining proteins exclusively identified in the fluconazole-sensitive strain were: Rcf1, Cef3, Pga43, Lcl3, Pfa5, and Tef1. No efflux pumps or key proteins in ergosterol biosynthesis were detected in this fluconazole-sensitive strain, thus implying the role of these proteins in conferring resistance to fluconazole.
In summary, this pilot study successfully detected CWPs expressed in a fluconazole-resistant strain upon exposure to fluconazole. Most of the cell wall proteins that were expressed and identified belong to pathways involved in resistance and pathogenicity in accordance with our previously observed phenotypes, a testament to the dynamic nature of cell wall protein expression and deposition in response to fluconazole stress. Some of these identified proteins can be targets of novel therapeutic strategies. Future work will expand on this study and investigate differential protein expression between resistant and sensitive strains to better elucidate the role that azoles play in modulating specific cell wall protein expression. Similar studies have previously been carried out in our lab to compare
C. albicans mutant with wild-type strains under filamentous and non-filamentous conditions [
21,
24,
57,
58,
59].