Immunomodulatory Effects of Pentoxifylline: Profiling Data Based on RAW 264.7 Cellular Signaling
Abstract
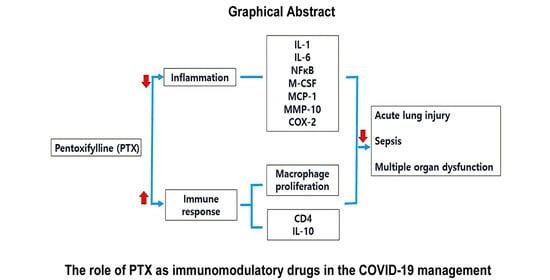
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Protein Extraction and IP-HPLC
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ward, A.; Clissold, S.P. Pentoxifylline. A review of its pharmacodynamic and pharmacokinetic properties, and its therapeutic efficacy. Drugs 1987, 34, 50–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xu, Y.J.; Mengi, S.A.; Ameja, A.S.; Dhalla, N.S. Therapeutic potentials of pentoxifylline for treatment of cardiovascular diseases. Exp. Clin. Cardiol. 2004, 9, 103–111. [Google Scholar] [PubMed]
- Magnusson, M.; Gunnarsson, M.; Berntorp, E.; Bjorkman, S.; Hoglund, P. Effects of pentoxifylline and its metabolites on platelet aggregation in whole blood from healthy humans. Eur. J. Pharmacol. 2008, 581, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Neuner, P.; Klosner, G.; Schauer, E.; Pourmojib, M.; Macheiner, W.; Grünwald, C.; Knobler, R.; Schwartz, A.; Luger, T.A.; Schwarz, T. Pentoxifylline in vivo down-regulates the release of IL-1 beta, IL-6, IL-8 and tumour necrosis factor-alpha by human peripheral blood mononuclear cells. Immunology 1994, 83, 262–267. [Google Scholar] [PubMed]
- Assimakopoulos, S.F.; Seintis, F.; Marangos, M. Pentoxifylline and complicated COVID-19: A pathophysiologically based treatment proposal. Med. Hypotheses 2020, 143, 109926. [Google Scholar] [CrossRef]
- Rübe, C.E.; Wilfert, F.; Uthe, D.; Schmid, K.W.; Knoop, R.; Willich, N.; Schuck, A.; Rübe, C. Modulation of radiation-induced tumour necrosis factor alpha (TNF-alpha) expression in the lung tissue by pentoxifylline. Radiother. Oncol. 2002, 64, 177–187. [Google Scholar] [CrossRef]
- Zein, C.O.; Yerian, L.M.; Gogate, P.; Lopez, R.; Kirwan, J.P.; Feldstein, A.E.; McCullough, A.J. Pentoxifylline improves nonalcoholic steatohepatitis: A randomized placebo-controlled trial. Hepatology 2011, 54, 1610–1619. [Google Scholar] [CrossRef]
- Speer, E.M.; Diago-Navarro, E.; Ozog, L.S.; Dowling, D.J.; Hou, W.; Raheel, M.; Fries, F.C.; Levy, O. Pentoxifylline alone or in combination with gentamicin or vancomycin inhibits live microbe-induced pro-inflammatory cytokine production in human cord blood and cord blood monocytes in vitro. Antimicrob. Agents Chemother. 2018, 62, e01462-18. [Google Scholar] [CrossRef] [Green Version]
- Shabaan, A.E.; Nasef, N.; Shouman, B.; Nour, I.; Mesbah, A.; Abdel-Hady, H. Pentoxifylline therapy for late-onset sepsis in preterm infants: A randomized controlled trial. Pediatr. Infect. Dis. J. 2015, 34, e143–e148. [Google Scholar] [CrossRef]
- Poggi, C.; Dani, C. Sepsis and oxidative stress in the newborn: From pathogenesis to novel therapeutic targets. Oxid. Med. Cell. Longev. 2018, 2018, 9390140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, D.M. Pentoxifylline inhibits the proliferation of human fibroblasts derived from keloid, scleroderma and morphoea skin and their production of collagen, glycosaminoglycans and fibronectin. Br. J. Dermatol. 1990, 123, 339–346. [Google Scholar] [CrossRef]
- Duncan, H.A.; Berman, B. Pentoxifylline and interferons decrease type I and III procollagen mRNA levels in dermal fibroblasts: Evedence for mediation by nuclear factor 1 down-regulation. J. Invest. Dermatol. 1995, 104, 282–286. [Google Scholar] [CrossRef] [Green Version]
- Horvath, B.; Marton, Z.; Halmosi, R.; Alexy, T.; Szapary, L.; Vekasi, J.; Biro, Z.; Habon, T.; Kesmarky, G.; Toth, K. In vitro antioxidant properties of pentoxifylline, piracetam, and vinpocetine. Clin. Neuropharmacol. 2002, 25, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Chen, R.H.; Chen, Y.M.; Chiang, W.C.; Lai, C.F.; Wu, K.D.; Tsai, T.J. Pentoxifylline attenuates tubulointerstitial fibrosis by blocking Smad3/4-activated transcription and profibrogenic effects of connective tissue growth factor. J. Am. Soc. Nephrol. 2005, 16, 2702–2713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boerma, M.; Roberto, K.A.; Hauer-Jensen, M. Prevention and treatment of functional and structural radiation injury in the rat heart by pentoxifylline and alpha-tocopherol. Int. J. Radiat. Oncol. Biol. Phys. 2008, 72, 170–177. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.L.; Lee, M.G.; Lee, C.C.; Su, P.I.; Chi, C.Y.; Liu, C.H.; Wu, M.C.; Yen, Z.S.; Chen, S.C. Pentoxifylline decreases post-operative intra-abdominal adhesion formation in an animal model. PeerJ 2018, 6, e5434. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Shim, S.; Kim, M.J.; Myung, J.K.; Jang, W.S.; Bae, C.H.; Lee, S.J.; Kim, M.K.; Jin, Y.W.; Lee, S.S.; et al. Pentoxifylline regulates plasminogen activator inhibitor-1 expression and protein kinase A phosphorylation in radiation-induced lung fibrosis. Biomed. Res. Int. 2017, 2017, 1279280. [Google Scholar] [CrossRef]
- O’Dell, K.; Sinha, U. Osteoradionecrosis. Oral Maxillofac. Surg. Clin. N. Am. 2011, 23, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Delanian, S.; Lefaix, J.L. The radiation-induced fibroatrophic process: Therapeutic perspective via the antioxidant pathway. Radiother. Oncol. 2004, 73, 119–131. [Google Scholar] [CrossRef]
- Seo, M.H.; Myoung, H.; Lee, J.H.; Yang, H.C.; Woo, K.M.; Lee, S.K.; Kim, S.M. Effects of pentoxifylline and tocopherol on an osteoradionecrosis animal model. J. Craniomaxillofac. Surg. 2020, 48, 621–631. [Google Scholar] [CrossRef]
- Delanian, S.; Porcher, R.; Balla-Mekias, S.; Lefaix, J.L. Randomized, placebo-controlled trial of combined pentoxifylline and tocopherol for regression of superficial radiation-induced fibrosis. J. Clin. Oncol. 2003, 21, 2545–2550. [Google Scholar] [CrossRef]
- Delanian, S.; Chatel, C.; Porcher, R.; Depondt, J.; Lefaix, J.L. Complete restoration of refractory mandibular osteoradionecrosis by prolonged treatment with a pentoxifylline-tocopherol-clodronate combination (PENTOCLO): A phase II trial. Int. J. Radiat. Oncol. Biol. Phys. 2011, 80, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Seirafianpour, F.; Mozafarpoor, S.; Fattahi, N.; Sadeghzadeh-Bazargan, A.; Hanifiha, M.; Goodarzi, A. Treatment of COVID-19 with pentoxifylline: Could it be a potential adjuvant therapy? Derm. Ther. 2020, 33, e13733. [Google Scholar] [CrossRef]
- Monji, F.; Al-Mahmood, S.A.; Hashemian, F. Can pentoxifylline and similar xanthine derivatives find a niche in COVID-19 therapeutic strategies? A ray of hope in the midst of the pandemic. Eur. J. Pharmacol. 2020, 887, 173561. [Google Scholar] [CrossRef]
- Maldonado, V.; Loza-Mejía, M.A.; Chávez-Alderete, J. Repositioning of pentoxifyllline as an immunomodulator and regulator of the renin-angiotensin system in the treatment of COVID-19. Med. Hypotheses 2020, 144, 109988. [Google Scholar] [CrossRef]
- DiNicolantonio, J.J.; Barroso-Aranda, J. Harnessing Adenosine A2A Receptors as a Strategy for Suppressing the Lung Inflammation and Thrombotic Complications of COVID-19: Potential of Pentoxifylline and Dipyridamole. Med. Hypotheses 2020, 143, 110051. [Google Scholar] [CrossRef]
- Delanian, S.; Depondt, J.; Lefaix, J.L. Major healing of refractory mandible osteoradionecrosis after treatment combining pentoxifylline and tocopherol: A phase II trial. Head Neck 2005, 27, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Hoefert, S.; Schmitz, I.; Weichert, F.; Gaspar, M.; Eufinger, H. Macrophage and bisphosphonate-related osteonecrosis of the jaw (BRONJ): Evidence of local immunosuppression of macrophages in contrast to other infectious jaw diseases. Clin. Oral Investig. 2015, 19, 497–508. [Google Scholar] [CrossRef]
- Merad, S.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S. Protein expression changes induced by cisplatin in an oral cancer cell line as determined by immunoprecipitation-based high performance liquid chromatography. Korean J. Oral Maxillofac. Pathol. 2015, 39, 567–582. [Google Scholar] [CrossRef]
- Kim, Y.S.; Lee, S.K. IP-HPLC Analysis of Human Salivary Protein Complexes. Kor. J. Oral Maxillofac. Pathol. 2015, 39, 615–622. [Google Scholar] [CrossRef]
- Kim, S.M.; Eo, M.Y.; Cho, Y.J.; Kim, Y.S.; Lee, S.K. Differential protein expression in the secretory fluids of maxillary sinusitis and maxillary retention cyst. Eur. Arch. Otorhinolaryngol. 2017, 274, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.S.; Kim, M.K.; Kim, Y.S.; Lee, S.K. In vitro protein expression changes in RAW 264.7 cells and HUVECs treated with dialyzed coffee extract by immunoprecipitation high performance liquid chromatography. Sci. Rep. 2018, 8, 13841. [Google Scholar] [CrossRef]
- Lee, S.S.; Kim, S.M.; Kim, Y.S.; Lee, S.K. Extensive protein expression changes induced by pamidronate in RAW 264.7 cells as determined by IP-HPLC. PeerJ 2020, 8, e9202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Bharadwaj, U.; Li, M.; Chen, C.; Yao, Q. Effects of pentoxifylline on differentiation, maturation, and function of human CD14+ monocyte-derived dendritic cells. J. Immunother. 2007, 30, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tam, W.F.; Hughes, C.C.; Rath, S.; Sen, R. c-Rel is a target of pentoxifylline-mediated inhibition of T lymphocyte activation. Immunity 1997, 6, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Matricardi, P.M.; Dal Negro, R.W.; Nisini, R. The first, holistic immunological model of COVID-19: Implications for prevention, diagnosis, and public health measures. Pediatr. Allergy Immunol. 2020, 31, 454–470. [Google Scholar] [CrossRef]
- Seyed Hosseini, E.; Riahi Kashani, N.; Nikzad, H.; Azadbakht, J.; Hassani Bafrani, H.; Haddad Kashani, H. The novel coronavirus Disease-2019 (COVID-19): Mechanism of action, detection and recent therapeutic strategies. Virology 2020, 551, 1–9. [Google Scholar] [CrossRef]
- Stasi, C.; Fallani, S.; Voller, F.; Silvestri, C. Treatment for COVID-19: An overview. Eur. J. Pharmacol. 2020, 889, 173644. [Google Scholar] [CrossRef] [PubMed]
- Cao, X. COVID-19: Immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020, 20, 269–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bermejo Martin, J.F.; Jimenez, J.L.; Muńoz-Fernández, A. Pentoxifylline and severe acute respiratory syndrome (SARS): A drug to be considered. Med. Sci. Monit. 2003, 9, SR29–SR34. [Google Scholar]
- Strutz, F.; Heeg, M.; Kochsiek, T.; Siemers, G.; Zeisberg, M.; Muller, G.A. Effects of pentoxifylline, pentifylline and gamma-interferon on proliferation, differentiation, and matrix synthesis of human renal fibroblasts. Nephrol. Dial. Transpl. 2000, 15, 1535–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okunieff, P.; Augustine, E.; Hicks, J.E.; Cornelison, T.L.; Altemus, R.M.; Naydich, B.G.; Ding, I.; Huser, A.K.; Abraham, E.H.; Smith, J.J.; et al. Pentoxifylline in the treatment of radiation-induced fibrosis. J. Clin. Oncol. 2004, 22, 2207–2213. [Google Scholar] [CrossRef]
- Rabbani, Z.N.; Mi, J.; Zhang, Y.; Delong, M.; Jackson, I.L.; Fleckenstein, K.; Salahuddin, F.K.; Zhang, X.; Clary, B.; Anscher, M.S.; et al. Hypoxia inducible factor 1alpha signaling in fractionated radiation-induced lung injury: Role of oxidative stress and tissue hypoxia. Radiat. Res. 2010, 173, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and functional exhaustion of T cells in patients with Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and Immunologic features in severe and moderate Coronavirus Disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedersen, S.F.; Ho, Y.C. SARS-CoV-2: A storm is raging. J. Clin. Investig. 2020, 130, 2202–2205. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, L.; Li, J.; Luo, M.; Shang, B. Pentoxifylline induces apoptosis of HepG2 cells by reducing reactive oxygen species production and activating the MAPK signaling. Life Sci. 2017, 183, 60–68. [Google Scholar] [CrossRef]
- Luo, M.; Dong, L.; Li, J.; Wang, Y.; Shang, B. Protective effects of pentoxifylline on acute liver injury induced by thioacetamide in rats. Int. J. Clin. Exp. Pathol. 2015, 8, 8990–8996. [Google Scholar] [PubMed]
- Armagan, I.; Bayram, D.; Candan, I.A.; Yigit, A.; Celik, E.; Armagan, H.H.; Uguz, A.C. Effects of pentoxifylline and alpha lipoic acid on methotrexate-induced damage in liver and kidney of rats. Environ. Toxicol. Pharmacol. 2015, 39, 1122–1131. [Google Scholar] [CrossRef]
- Golunski, G.; Woziwodzka, A.; Piosik, J. Potential use of Pentoxifylline in cancer therapy. Curr. Pharm. Biotechnol. 2018, 19, 206–216. [Google Scholar] [CrossRef]
- Horiuchi, H.; Saito, N.; Kinoshita, T.; Wakabayashi, S.; Tsutsumimoto, T.; Otsuru, S.; Takaoka, K. Enhancement of recombinant human bone morphogenetic protein-2 (rhBMP-2)-induced new bone formation by concurrent treatment with parathyroid hormone and a phosphodiesterase inhibitor, pentoxifylline. J. Bone Miner. Metab. 2004, 22, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Cakmak, G.; Sahin, M.S.; OzdemIr, B.H.; Karadeniz, E. Effect of pentoxifylline on healing of segmental bone defects and angiogenesis. Acta Orthop. Traumatol. Turc. 2016, 49, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Kamran, M.Z.; Gude, R.P. Pentoxifylline inhibits melanoma tumor growth and angiogenesis by targeting STAT3 signaling pathway. Biomed. Pharmacother. 2013, 67, 399–405. [Google Scholar] [CrossRef] [PubMed]









| Signaling Proteins | No. | Antibodies |
|---|---|---|
| Cellular proliferation | 10 | Ki-67 *, PCNA *, CDK4 *, PLK4 *, MPM2 *, p14 *, p16 *, p21 *, p27 *, cyclin D2 * |
| cMyc/MAX/MAD signaling | 3 | cMyc *, MAX *, MAD * |
| p53/Rb/E2F signaling | 4 (2) | p53, Rb-1 #, E2F-1 *, MDM2 *, CDK4 *, p21 * |
| Epigenetic modification | 6 | DMAP1, histone H1 *, KDM4D $, HDAC-10 $, MBD4, DNMT1 |
| Protein translation signaling | 3 | DOHH *, DHS *, elF5A-1 $ |
| RAS signaling | 7 (3) | NRAS $, KRAS $, HRAS, Rab, JNK-1 *, ERK-1 *, MEKK, (pAKT1/2/3 *, mTOR, PKC) |
| Growth factor signaling | 9 | TGF-β1 #, IGF-1 *, HER1 *, HER2 *, ERβ *, FGF-1, FGF-2, Met, CTGF |
| NFkB signaling | 9 (3) | NFkB *, IKK *, NFATS *, PGC-1α, GADD45 *, GADD153 *, mTOR @, p38 *, MDR *, (ERK-1 *, pAKT1/2/3 *, TNFα *) |
| Immunity signaling | 7 | CD3 *, CD4, CD20 *, CD28 *, CD31 *, CD68 *, cathepsin K * |
| Inflammatory signaling | 20 | TNFα @, IL-1 *, IL-6 *, IL-10 *, IL-28 *, COX-2 *, lysozyme *, M-CSF *, MMP-1 *, MMP-2 *, MMP-3 *, MMP-9 *, MMP-10 *, MMP-12, MCP-1, LTA4H, CXCR4, TLR3, hepcidin, CRP-1 * |
| Cell protection | 8 (3) | HSP-70 *, HSP-90 *, AP-1 *, SP-1 *, SP-3 *, p38 *, PKC *, pAKT1/2/3 *, (p38, JNK-1, TERT) |
| Cellular differentiation | 6 (4) | PLC-β2, TGase-2, HXKII *, Jagged-2, Notch-1, GLI-1, (PKC, AP-1, SP-1, SP-3) |
| Antioxidant-related proteins | 5 | SOD-1 *, GST *, LC3 *, AMPK *, NOS-1, |
| p53-mediated cellular apoptosis | 6 (1) | p53 *, MDM2 *, BAX *, BAK *, APAF-1 *, caspase 9 *, (PARP) |
| FAS-mediated cellular apoptosis | 6 | FASL *, FAS *, FADD *, FLIP *, caspase 8 *, PARP * |
| Oncogenic proteins | 3 (3) | 14-3-3 *, TERT *, YAP, (pAKT1/2/3, MBD4, mTOR) |
| Angiogenesis-related proteins | 8 (4) | HIF &, VEGF-A *, VEGF-C *, VCAM, angiogenin *, CMG2 $, vWF $, ET-1 *(CD31, MMP-2, MMP-10, FGF-2) |
| Osteogenesis-related proteins | 9 (2) | RANKL, OPG *, osteonectin, osteopontin, osteocalcin, RUNX2, ALP *, osterix *, BMP-2 * (cathepsin K, HSP-90) |
| Control cytoplasmic proteins | 3 | α-tubulin *, β-actin *, GAPDH * |
| Total | 132 (25) |
| Signaling | Up-Regulated | Unchanged | Down-Regulated |
|---|---|---|---|
| Proliferation ↓ | PCNA | Ki-67, CDK4, PLK4, MPM2, cyclin D2, Rb-1, E2F-1, MDM2 | |
| Apoptosis ↓ | BAX, APAF-1 | BAK, caspase 9 | p53, PARP, FASL, FADD, caspase 8 |
| Antioxidant and cellular protection | AP-1, SP-1, AMPK-1 | NOS-1, SOD-1, GST | |
| Angiogenesis | HIF, angiogenin, VEGF-C | VEGF-A, vWF, ET-1, CD31 | CMG2 |
| Osteogenesis ↑ | OPG, RANKL, osteopontin, RUNX2, osterix | osteonectin, ALP | Cathepsin K |
| Inflammation ↓ | CD4, IL-10, MMP-1, MMP-2, MMP-3, MMP-9, | TLR3, CD31, CD68, lysozyme, TNF-α | IL-1, IL-6, CD3, CD20, CD28, M-CSF, cathepsin K, LTA4H, CXCR4, COX-2 |
| Growth factor | IGF-1, HER1, HER2 | TGF-β1 | FGF-1, FGF-2, ERβ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, M.H.; Eo, M.Y.; Nguyen, T.T.H.; Yang, H.J.; Kim, S.M. Immunomodulatory Effects of Pentoxifylline: Profiling Data Based on RAW 264.7 Cellular Signaling. Appl. Sci. 2021, 11, 8273. https://0-doi-org.brum.beds.ac.uk/10.3390/app11178273
Seo MH, Eo MY, Nguyen TTH, Yang HJ, Kim SM. Immunomodulatory Effects of Pentoxifylline: Profiling Data Based on RAW 264.7 Cellular Signaling. Applied Sciences. 2021; 11(17):8273. https://0-doi-org.brum.beds.ac.uk/10.3390/app11178273
Chicago/Turabian StyleSeo, Mi Hyun, Mi Young Eo, Truc Thi Hoang Nguyen, Hoon Joo Yang, and Soung Min Kim. 2021. "Immunomodulatory Effects of Pentoxifylline: Profiling Data Based on RAW 264.7 Cellular Signaling" Applied Sciences 11, no. 17: 8273. https://0-doi-org.brum.beds.ac.uk/10.3390/app11178273