Comparative Interactions of Dihydroquinazolin Derivatives with Human Serum Albumin Observed via Multiple Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Apparatus
2.2. Reagents
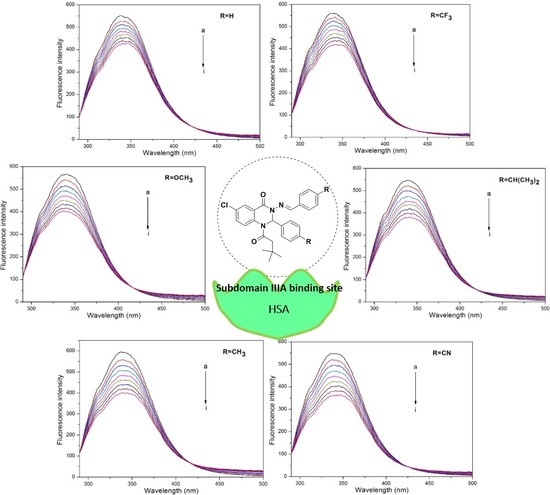
2.3. Fluorescence Spectra Measurements
2.4. Fluorescence Titration Experiments
2.5. Site Marker Competitive Replacement Study
2.6. Circular Dichroism Spectra Studies
2.7. Fourier Transform Infrared Measurements
2.8. Synthesis of 6-Chloro-1-(3,3-dimethyl-butanoyl)-2-(un)substituted-phenyl-2,3-dihydroquinazolin-4(1H)-one Derivatives 3a–f
3. Results and Discussion
3.1. Synthesis
3.2. Fluorescence Quenching Mechanism
3.3. Binding Sites and Identification of Binding Sites on HSA
3.4. Thermodynamic Parameters and Binding Modes
3.5. HSA Conformational Change Evaluated Using CD and FTIR Measurements
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Alam, P.; Abdelhameed, A.S.; Rajpoot, R.K.; Khan, R.H. Interplay of Multiple Interaction Forces: Binding of Tyrosine Kinase Inhibitor Nintedanib with Human Serum Albumin. J. Photochem. Photobiol. B 2016, 157, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.R.; Khan, S.I.; Singh, S.; Khan, I.A.; Vishwakarma, R.A.; Bharate, S.B. Synthesis, Antimalarial and Antitubercular Activities of Meridianin Derivatives. Eur. J. Med. Chem. 2015, 98, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Yousef, T.A.; Alduaij, O.K.; Abu El-Reash, G.M.; El Morshedy, R.M. Semiempirical Studies, Spectral Analysis, in vitro Antibacterial and DNA Degradation Studies of Heterocyclic Thiosemicarbazone Ligand and Its Metal Complexes. J. Mol. Liq. 2016, 222, 762–776. [Google Scholar] [CrossRef]
- Hemalatha, K.; Madhumitha, G.; Ravi, L.; Khanna, V.G.; Al-Dhabi, N.A.; Arasu, M.V. Binding Mode of Dihydroquinazolinones with Lysozyme and Its antifungal Activity Against Aspergillus Species. J. Photochem. Photobiol. B 2016, 161, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, M.; Yao, X.; Li, Y.; Tan, J.; Wang, L.; Qiao, W.; Geng, Y.; Liu, Y.; Wang, Q. Design, Synthesis and Antiviral Activity of Novel Quinazolinones. Eur. J. Med. Chem. 2012, 53, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, A.A.M.; Abou-Zeid, L.A.; ElTahir, K.E.H.; Mohamed, M.A.; Abu El-Enin, M.A.; El-Azab, A.S. Design, Synthesis of 2,3-disubstituted 4(3H)-quinazolinone Derivatives as Anti-inflammatory and Analgesic Agents: COX-1/2 Inhibitory Activities and Molecular Docking Studies. Bioorg. Med. Chem. 2016, 24, 3818–3828. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, A.A.M.; Abou-Zeid, L.A.; El Tahir, K.E.H.; Ayyad, R.R.; El-Sayed, M.A.A.; El-Azab, A.S. Synthesis, Anti-inflammatory, Analgesic, COX-1/2 Inhibitory Activities and Molecular Docking Studies of Substituted 2-mercapto-4(3H)-quinazolinones. Eur. J. Med. Chem. 2016, 121, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Georgey, H.; Abdel-Gawad, N.; Abbas, S. Synthesis and Anticonvulsant Activity of Some Quinazolin-4-(3H)-one Derivatives. Molecules 2008, 13, 2557–2569. [Google Scholar] [CrossRef] [PubMed]
- Khattab, S.N.; Haiba, N.S.; Asal, A.M.; Bekhit, A.A.; Amer, A.; Abdel-Rahman, H.M.; El-Faham, A. Synthesis and Evaluation of Quinazoline Amino acid Derivatives as Mono Amine Oxidase (MAO) Inhibitors. Bioorg. Med. Chem. 2015, 23, 3574–3585. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Polovka, M.; Martinez-Ayala, A.L.; González-Aguilar, G.A.; Ham, K.S.; Kang, S.G.; Park, Y.K.; Heo, B.G.; Namiesnik, J.; Gorinstein, S. Fluorescence Studies by Quenching and Protein Unfolding on the Interaction of Bioactive Compounds in Water Extracts of Kiwi Fruit Cultivars with Human Serum Albumin. J. Lumin. 2015, 160, 71–77. [Google Scholar] [CrossRef]
- Saeidifar, M.; Mansouri-Torshizi, H.; Saboury, A.A. Biophysical Study on the Interaction between two Palladium(II) Complexes and Human Serum Albumin by Multispectroscopic Methods. J. Lumin. 2015, 167, 391–398. [Google Scholar] [CrossRef]
- Sekar, G.; Sugumar, S.; Mukherjee, A.; Chandrasekaran, N. Multiple Spectroscopic Studies of the Structural Conformational Changes of Human Serum Albumin-Essential Oil Based Nanoemulsions Conjugates. J. Lumin. 2015, 161, 187–197. [Google Scholar] [CrossRef]
- Zhu, G.; Wang, Y.; Xi, L.; Liu, J.; Wang, H.; Du, L. Spectroscopy and Molecular Docking Studies on the Binding of Propyl Gallate to Human Serum Albumin. J. Lumin. 2015, 159, 188–196. [Google Scholar] [CrossRef]
- Galecki, K.; Hunter, K.; Dankova, G.; Rivera, E.; Tung, L.W.; McSherry, K. Experimental and Theoretical Investigation of Bezafibrate Binding to Serum Albumins. J. Lumin. 2016, 177, 235–241. [Google Scholar] [CrossRef]
- Ding, F.; Zhao, G.; Chen, S.; Liu, F.; Sun, Y.; Zhang, L. Chloramphenicol Binding to Human Serum Albumin: Determination of Binding Constants and Binding Sites by Steady-state Fluorescence. J. Mol. Struct. 2009, 929, 159–166. [Google Scholar] [CrossRef]
- Macmanusspencer, L.A.; Tse, M.L.; Hebert, P.C.; Bischel, H.N.; Luthy, R.G. Binding of Perfluorocarboxylates to Serum Albumin: A Comparison of Analytical Methods. Anal. Chem. 2010, 82, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Diao, J.; Sun, Y.; Sun, Y. Bioevaluation of Human Serum Albumin-Hesperidin Bioconjugate: Insight into Protein Vector Function and Conformation. J. Agric. Food Chem. 2012, 60, 7218–7228. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Liu, W.; Li, Y.; Zhang, L.; Sun, Y. Determining the Binding Affinity and Binding Site of Bensulfuron-methyl to Human Serum Albumin by Quenching of the Intrinsic Tryptophan Fluorescence. J. Lumin. 2010, 130, 2013–2021. [Google Scholar] [CrossRef]
- Huang, S.; Qiu, H.; Lu, S.; Zhu, F.; Xiao, Q. Study on the Molecular Interaction of Graphene Quantum Dots with Human Serum Albumin: Combined Spectroscopic and Electrochemical Approaches. J. Hazard. Mater. 2015, 285, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Diao, J.; Yang, X.; Sun, Y. Structural Analysis and Binding Domain of Albumin Complexes with Natural Dietary Supplement Humic Acid. J. Lumin. 2011, 131, 2244–2251. [Google Scholar] [CrossRef]
- Pérez, D.I.; Pistolozzi, M.; Palomo, V.; Redondo, M.; Fortugno, C.; Gil, C.; Felix, G.; Martinez, A.; Bertucci, C. 5-Imino-1,2-4-thiadiazoles and Quinazolines Derivatives as Glycogen Synthase Kinase 3β (GSK-3β) and Phosphodiesterase 7 (PDE7) Inhibitors: Determination of Blood–brain Barrier Penetration and Binding to Human Serum Albumin. Eur. J. Pharm. Sci. 2012, 45, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, Y.; Lv, C.; Wang, L.; Ou, J.; Wang, M.; Liu, S. Impact of Halogen Substituents on Interactions between 2-Phenyl-2,3-dihydroqulinazolin-4(1H)-one Derivatives and Human Serum Albumin. Molecules 2012, 17, 2000–2014. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, M.; Liu, F.; Wu, X.; Pan, D.; Liu, J.; Fan, S.; Wang, Z.; Tang, J.; Na, R.; Li, Q.L.; Hua, R.; Liu, S. Comparative Studies of Interactions between Fluorodihydroquinazolin Derivatives and Human Serum Albumin with Fluorescence Spectroscopy. Molecules 2016, 21, 1373–1386. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 2015, 72, 248–254. [Google Scholar] [CrossRef]
- Ding, F.; Liu, W.; Diao, J.; Sun, Y. Characterization of Alizarin Red S binding sites and structural changes on human serum albumin: A biophysical study. J. Hazard. Mater. 2011, 186, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Sircar, J.C.; Capiris, T.; Kesten, S.J.; Herzig, D.J. Pyrazolo[5,1-b]quinazolin-9-ones: A New Series of Antiallergic agents. J. Med. Chem. 1981, 24, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Fulop, F.; Simeonov, M.; Pihlaja, K. Formation of 1,2-dihydroquinazolin-4(3H)-ones. Reinvestigation of a Recently Reported 1,3,4-benzotriazepine synthesis. Tetrahedron 1992, 48, 531–538. [Google Scholar] [CrossRef]
- Bi, S.; Zhao, T.; Zhou, H.; Wang, Y.; Li, Z. Probing the Interactions of Bromchlorbuterol-HCl and Phenylethanolamine A with HSA by Multi-spectroscopic and Molecular Docking Technique. J. Chem. Thermodyn. 2016, 97, 113–121. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Green, R.J.; Frazier, R.A. Interaction of Flavonoids with Bovine Serum Albumin: A Fluorescence Quenching Study. J. Agric. Food Chem. 2005, 53, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.N.; Islam, B.; Yennamalli, R.; Sultan, A.; Subbarao, N.; Khan, A.U. Interaction of Mitoxantrone with Human Serum Albumin: Spectroscopic and Molecular Modeling Studies. Eur. J. Pharm. Sci. 2008, 35, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Chen, Q. The Interaction between Ionic Liquids Modified Magnetic Nanoparticles and Bovine Serum Albumin and the Cytotoxicity to HepG-2 cells. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 120, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Mechanisms and Dynamics of Fluorescence Quenching. In Principles of Fluorescence Spectroscopy; Springer: Boston, MA, USA, 2006; pp. 331–351. [Google Scholar]
- Lakowicz, J.R.; Weber, G. Quenching of Fluorescence by Oxygen. Probe for Structural Fluctuations in Macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Cacita, N.; Nikolaou, S. Studying the Interaction between Trinuclear Ruthenium Complexes and Human Serum Albumin by Means of Fluorescence Quenching. J. Lumin. 2016, 169, 115–120. [Google Scholar] [CrossRef]
- Ghorab, M.M.; Alsaid, M.S.; Al-Dosari, M.S.; El-Gazzar, M.G.; Parvez, M.K. Design, Synthesis and Anticancer Evaluation of Novel Quinazoline-sulfonamide Hybrids. Molecules 2016, 21, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Zhang, L.; Sun, Y.; Diao, J.-X.; Yang, X.-L.; Sun, Y.; Zhang, L. Specificity and Affinity of Phenosafranine Protein Adduct: Insights from Biophysical Aspects. J. Lumin. 2012, 132, 629–635. [Google Scholar] [CrossRef]
- Trivedi, V.D.; Vorum, H.; Honore, B.; Qasim, M.A. Molecular Basis of Indomethacin-human Serum Albumin Interaction. J. Pharm. Pharmacol. 1999, 51, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Fan, J.; Li, J.; Hu, Z. Interactions between 1-benzoyl-4-p-chlorophenyl Thiosemicarbazide and Serum Albumin: Investigation by Fluorescence Spectroscopy. Bioorg. Med. Chem. 2004, 12, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Qin, Y.; Lin, A.; Hu, X.; Zou, G. Interaction of Daunomycin Antibiotic with Human Serum Albumin: Investigation by Resonant Mirror Biosensor Technique, Fluorescence Spectroscopy and Molecular Modeling Methods. J. Pharm. Biomed. Anal. 2005, 39, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Wang, Y.; Li, Z.; Dong, C. Fluorescence Study on the Interaction of Human Serum Albumin with Bromsulphalein. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2006, 65, 1144–1147. [Google Scholar] [CrossRef] [PubMed]
- Sjöholm, I.; Ekman, B.; Kober, A.; Ljungstedt-Påhlman, I.; Seiving, B.; Sjödin, T. Binding of Drugs to Human Serum Albumin:XI. The Specificity of Three Binding Sites as Studied with Albumin Immobilized in Microparticles. Mol. Pharmacol. 1979, 16, 767–777. [Google Scholar] [PubMed]
- Leckband, D. Measuring the Forces that Control Protein Interactions. Annu. Rev. Biophys. Biomol. Struct. 2003, 29, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.D.; Subramanian, S. Thermodynamics of Protein Association Reactions: Forces Contributing to Stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]






| Compound | T (K) | KSV (×104 M−1) | Kq (×1012 M−1·s−1) | R a | SD b |
|---|---|---|---|---|---|
| R = H | 298 | 2.769 | 2.769 | 0.99956 | 0.00322 |
| 307 | 2.976 | 2.976 | 0.99933 | 0.00424 | |
| 316 | 3.268 | 3.268 | 0.99919 | 0.00513 | |
| R = CH3 | 298 | 4.228 | 4.228 | 0.99942 | 0.00562 |
| 307 | 4.337 | 4.337 | 0.99924 | 0.00660 | |
| 316 | 4.513 | 4.513 | 0.99903 | 0.00776 | |
| R = OCH3 | 298 | 4.062 | 4.062 | 0.99948 | 0.00512 |
| 307 | 4.280 | 4.280 | 0.99889 | 0.00788 | |
| 316 | 4.478 | 4.478 | 0.99869 | 0.00896 | |
| R = CN | 298 | 4.961 | 4.961 | 0.99934 | 0.00705 |
| 307 | 5.088 | 5.088 | 0.99847 | 0.01100 | |
| 316 | 5.214 | 5.214 | 0.99824 | 0.01209 | |
| R = CF3 | 298 | 3.436 | 3.436 | 0.99928 | 0.00511 |
| 307 | 3.507 | 3.507 | 0.99921 | 0.00545 | |
| 316 | 3.623 | 3.623 | 0.99892 | 0.00658 | |
| R = CH(CH3)2 | 298 | 4.326 | 4.326 | 0.99939 | 0.00589 |
| 307 | 4.423 | 4.423 | 0.99894 | 0.00795 | |
| 316 | 4.593 | 4.593 | 0.99852 | 0.00975 |
| Compound | T (K) | Ka (×104 M−1) | n | R a | SD b |
|---|---|---|---|---|---|
| R = H | 298 | 2.15794 | 0.99314 | 0.99945 | 0.01088 |
| 307 | 2.34365 | 0.98423 | 0.99947 | 0.01059 | |
| 316 | 2.54637 | 0.97492 | 0.99928 | 0.01220 | |
| R = CF3 | 298 | 3.52395 | 1.03096 | 0.99959 | 0.00978 |
| 307 | 3.72392 | 1.02821 | 0.99966 | 0.00884 | |
| 316 | 3.90369 | 1.04261 | 0.99953 | 0.01056 | |
| R = OCH3 | 298 | 3.57039 | 1.00301 | 0.99949 | 0.01055 |
| 307 | 3.79393 | 1.01549 | 0.99918 | 0.01354 | |
| 316 | 4.06275 | 1.01981 | 0.99909 | 0.01435 | |
| R = CH(CH3)2 | 298 | 5.16790 | 1.05812 | 0.99965 | 0.00921 |
| 307 | 5.31304 | 1.06299 | 0.99961 | 0.00976 | |
| 316 | 5.46879 | 1.06713 | 0.99947 | 0.01149 | |
| R = CH3 | 298 | 3.58307 | 0.99055 | 0.99943 | 0.01102 |
| 307 | 3.69690 | 0.99108 | 0.99937 | 0.01160 | |
| 316 | 3.78131 | 0.99673 | 0.99895 | 0.01511 | |
| R = CN | 298 | 4.79585 | 1.02042 | 0.99959 | 0.00962 |
| 307 | 4.95109 | 1.03849 | 0.99923 | 0.01343 | |
| 316 | 5.08399 | 1.04716 | 0.99896 | 0.01578 |
| Compound | Site Marker | Ka (×104 M−1) | R a | SD b |
|---|---|---|---|---|
| R = H | Blank | 2.15794 | 0.99949 | 0.26651 |
| PB | 1.87645 | 0.99962 | 0.2702 | |
| FA | 1.04135 | 0.99948 | 0.25962 | |
| Dig | 2.17623 | 0.99938 | 0.24163 | |
| R = CF3 | Blank | 3.52395 | 0.99985 | 0.12641 |
| PB | 3.44165 | 0.99971 | 0.18126 | |
| FA | 3.03507 | 0.99985 | 0.18132 | |
| Dig | 4.02741 | 0.99976 | 0.15165 | |
| R = OCH3 | Blank | 3.57039 | 0.99965 | 0.15519 |
| PB | 2.89855 | 0.99941 | 0.22308 | |
| FA | 2.45095 | 0.99977 | 0.15596 | |
| Dig | 3.72478 | 0.99936 | 0.20352 | |
| R = CH(CH3)2 | Blank | 5.16790 | 0.99977 | 0.13434 |
| PB | 4.50395 | 0.99994 | 0.07291 | |
| FA | 2.91938 | 0.99953 | 0.22246 | |
| Dig | 5.08774 | 0.99995 | 0.05945 | |
| R = CH3 | Blank | 3.58307 | 0.99954 | 0.16563 |
| PB | 3.43302 | 0.99964 | 0.16227 | |
| FA | 2.95264 | 0.99987 | 0.11889 | |
| Dig | 3.75253 | 0.99943 | 0.18557 | |
| R = CN | Blank | 4.79585 | 0.99987 | 0.08076 |
| PB | 4.79839 | 0.99990 | 0.07751 | |
| FA | 3.28633 | 0.99934 | 0.19871 | |
| Dig | 5.00248 | 0.99952 | 0.14806 |
| Compound | Van ’t Hoff | R a | SD b |
|---|---|---|---|
| R=H | Y = 12.88527 − 866.16/T | 0.99975 | 0.00264 |
| R=CF3 | Y = 12.26971 − 536.08/T | 0.99971 | 0.00175 |
| R=OCH3 | Y = 12.74821 − 675.57/T | 0.99845 | 0.00509 |
| R = CH(CH3)2 | Y = 11.84592 − 296.08/T | 0.99944 | 0.00133 |
| R=CH3 | Y = 11.43527 − 282.37/T | 0.99742 | 0.00274 |
| R=CN | Y = 11.80447 − 305.68/T | 0.99949 | 0.00132 |
| Compound | T (K) | Ka (×104 M−1) | R a | ΔH (kJ·mol−1) | ΔG (kJ·mol−1) | ΔS (Jmol−1·K−1) |
|---|---|---|---|---|---|---|
| R = H | 298 | 2.15794 | 0.99949 | 7.20125 | −24.7229 | 107.1281 |
| 307 | 2.34365 | 0.99969 | −25.6871 | |||
| 316 | 2.54637 | 0.99963 | −26.6513 | |||
| R = CF3 | 298 | 3.52395 | 0.99985 | 4.45697 | −25.9421 | 102.0104 |
| 307 | 3.72392 | 0.99994 | −26.8602 | |||
| 316 | 3.90369 | 0.99980 | −27.7783 | |||
| R = OCH3 | 298 | 3.57039 | 0.99965 | 5.61669 | −25.9679 | 105.9886 |
| 307 | 3.79393 | 0.99944 | −26.9218 | |||
| 316 | 4.06275 | 0.99961 | −27.8757 | |||
| R = CH(CH3)2 | 298 | 5.16790 | 0.99977 | 2.41609 | −26.9330 | 98.4869 |
| 307 | 5.31304 | 0.99984 | −27.8194 | |||
| 316 | 5.46879 | 0.99990 | −28.7058 | |||
| R=CH3 | 298 | 3.58307 | 0.99954 | 2.34762 | −25.9841 | 95.0728 |
| 307 | 3.69690 | 0.99958 | −26.8397 | |||
| 316 | 3.78131 | 0.99925 | −27.6954 | |||
| R=CN | 298 | 4.79585 | 0.99987 | 2.54142 | −26.7050 | 98.1424 |
| 307 | 4.95109 | 0.99968 | −27.5883 | |||
| 316 | 5.08399 | 0.99959 | −28.4716 |
| Compound | ΔΔH (kJ·mol−1) | ΔΔS (Jmol−1·K−1) |
|---|---|---|
| R = CF3 | −2.74428 | −5.1177 |
| R = OCH3 | −1.58456 | −1.1395 |
| R = CH(CH3)2 | −4.78516 | −8.6412 |
| R = CH3 | −4.85363 | −12.0553 |
| R = CN | −4.65983 | −8.9857 |
| T (K) | ΔΔG (kJ·mol−1) | ||||
|---|---|---|---|---|---|
| R = CF3 | R = OCH3 | R = CH(CH3)2 | R = CH3 | R = CN | |
| 298 | −1.2192 | −1.2450 | −2.2101 | −1.2612 | −1.9821 |
| 307 | −1.1731 | −1.2347 | −2.1323 | −1.1526 | −1.9012 |
| 316 | −1.1270 | −1.2226 | −2.0527 | −1.0441 | −1.8203 |
| Sample | Secondary Structure (%) | |||
|---|---|---|---|---|
| α-Helix | β-Sheet | β-Turn | Random Coil | |
| HSA | 38.8 | 23.6 | 11.0 | 26.6 |
| HSA + 3a (1:2) | 35.8 | 22.7 | 14.2 | 27.2 |
| HSA + 3b (1:2) | 37.8 | 24.7 | 10.7 | 26.8 |
| HSA + 3c (1:2) | 36.4 | 24.4 | 11.1 | 28.1 |
| HSA + 3d (1:2) | 36.4 | 20.0 | 14.8 | 28.8 |
| HSA + 3e (1:2) | 36.8 | 22.2 | 12.4 | 28.5 |
| HSA + 3f (1:2) | 33.7 | 14.5 | 19.5 | 32.2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhu, M.; Liu, J.; Na, R.; Liu, F.; Wu, X.; Fan, S.; Wang, Z.; Pan, D.; Tang, J.; et al. Comparative Interactions of Dihydroquinazolin Derivatives with Human Serum Albumin Observed via Multiple Spectroscopy. Appl. Sci. 2017, 7, 200. https://0-doi-org.brum.beds.ac.uk/10.3390/app7020200
Wang Y, Zhu M, Liu J, Na R, Liu F, Wu X, Fan S, Wang Z, Pan D, Tang J, et al. Comparative Interactions of Dihydroquinazolin Derivatives with Human Serum Albumin Observed via Multiple Spectroscopy. Applied Sciences. 2017; 7(2):200. https://0-doi-org.brum.beds.ac.uk/10.3390/app7020200
Chicago/Turabian StyleWang, Yi, Meiqing Zhu, Jia Liu, Risong Na, Feng Liu, Xiangwei Wu, Shisuo Fan, Zhen Wang, Dandan Pan, Jun Tang, and et al. 2017. "Comparative Interactions of Dihydroquinazolin Derivatives with Human Serum Albumin Observed via Multiple Spectroscopy" Applied Sciences 7, no. 2: 200. https://0-doi-org.brum.beds.ac.uk/10.3390/app7020200