Enhancing Interpretability of Gene Signatures with Prior Biological Knowledge
Abstract
:1. Introduction
2. Experimental Section
2.1. Data and Preprocessing
2.2. Methods
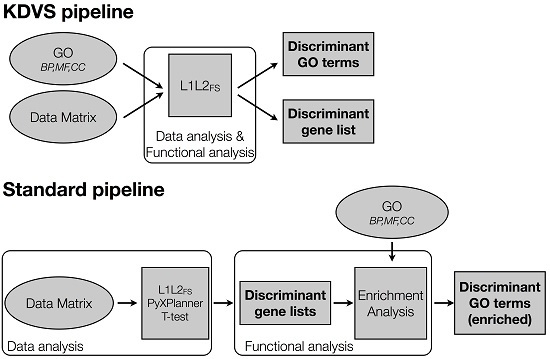
2.2.1. Experimental Framework
2.2.2. The Standard Pipeline
Data Analysis
Functional Analysis
2.2.3. The KDVS Pipeline
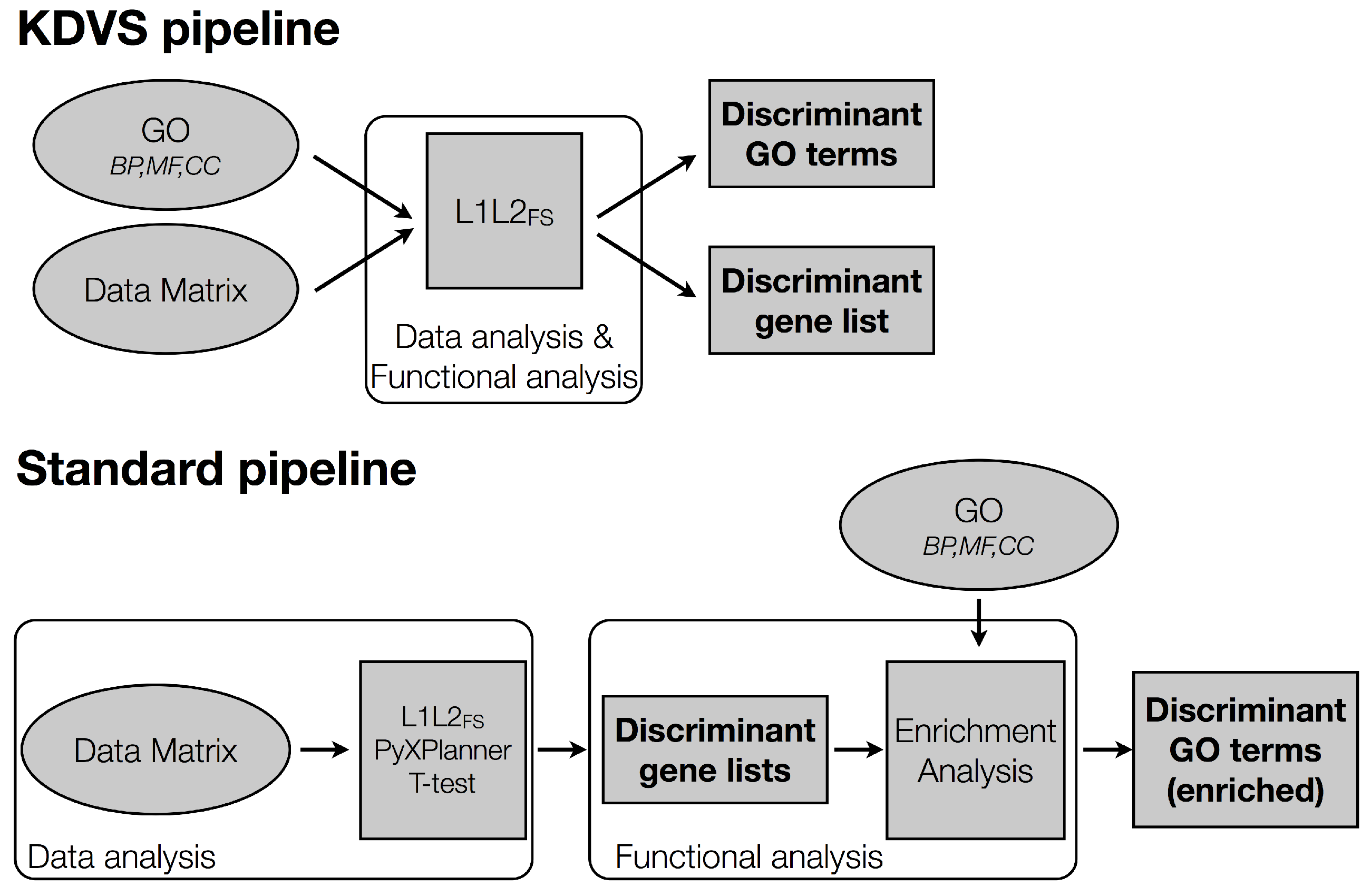
2.2.4. Benchmark Lists
2.2.5. Performance Metrics
3. Results
4. Discussion
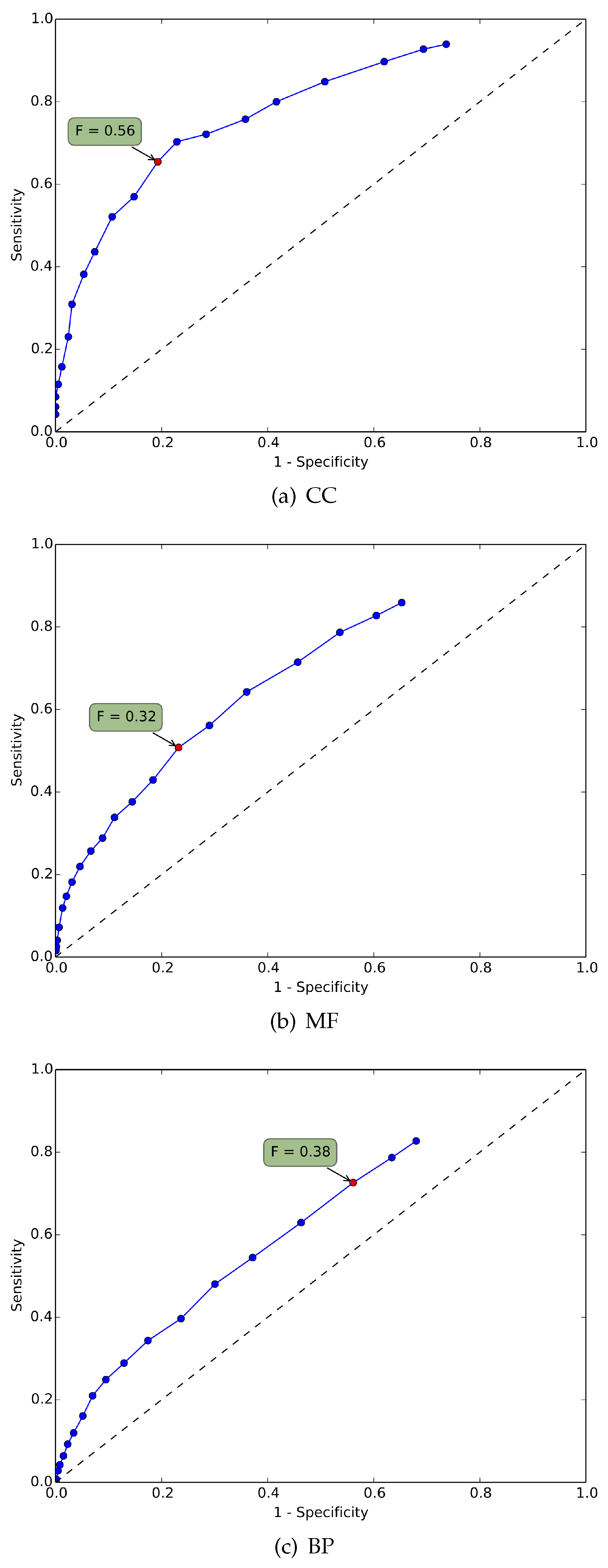
4.1. Statistical Analysis
4.2. Biological Significance
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PD | Parkinson’s Disease |
| GEO | Gene Expression Omnibus |
| KDVS | Knowledge Driven Variable Selection |
feature selection framework | |
| GO | Gene Ontology |
| FiltKBest | Filter K-Best selection based on ANOVA |
| LASSO | Lasso |
| ENET | Elastic Net |
| KNN | K Nearest Neighbor |
| LR | Logistic Regression |
| LSVM | Linear Support Vector Machine |
| OLS | Ordinary Least Square |
| RLS | regularized least square |
| DAG | Directed Acyclic Graph |
| MF | Molecular Function |
| BP | Biological Process |
| CC | Cellular Component |
| KEGG | Kyoto Encyclopaedia of Genes and Genomes |
| GOA | Gene Ontology Annotations |
| TAS | Traceable Author Statement |
| IC | Inferred by Curator |
| TP | True Positive |
| TN | True Negative |
| FP | False Positive |
| FN | False Negative |
References
- Huang, D.; Sherman, B.; Lempicki, R. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.; Blake, J.; Botstein, D.; Butler, H.; Cherry, J.; Davis, A.; Dolinski, K.; Dwight, S.; Eppig, J.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Zycinski, G.; Barla, A.; Squillario, M.; Sanavia, T.; Di Camillo, B.; Verri, A. Knowledge Driven Variable Selection (KDVS)—A new approach to enrichment analysis of gene signatures obtained from high-throughput data. Source Code Biol. Med. 2013, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wulf, A.; Liu, T.; Khoury, M.; Gwinn, M. Gene Prospector: An evidence gateway for evaluating potential susceptibility genes and interacting risk factors for human diseases. BMC Bioinform. 2008, 9, 528. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Annotations Documentation. Available online: ftp://ftp.geneontology.org/go/www/GO.gettingStarted.shtml (accessed on 3 June 2016).
- Edgar, R.; Domrachev, M.; Lash, A. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Lesnick, T.G.; Papapetropoulos, S.; Mash, D.C.; Ffrench-Mullen, J.; Shehadeh, L.; de Andrade, M.; Henley, J.R.; Rocca, W.A.; Ahlskog, J.E.; Maraganore, D.M. A Genomic Pathway Approach to a Complex Disease: Axon Guidance and Parkinson Disease. PLoS Genet. 2007, 3, e98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; James, M.; Middleton, F.A.; Davis, R.L. Transcriptional analysis of multiple brain regions in Parkinson’s disease supports the involvement of specific protein processing, energy metabolism, and signaling pathways, and suggests novel disease mechanisms. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2005, 137B, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Liao, Z.; Locascio, J.J.; Lesniak, K.A.; Roderick, S.S.; Watt, M.L.; Eklund, A.C.; Zhang-James, Y.; Kim, P.D.; Hauser, M.A.; et al. PGC-1α, A Potential Therapeutic Target for Early Intervention in Parkinson’s Disease. Sci. Transl. Med. 2010, 2, 52ra73–52ra73. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.; Bolstad, B.; Collin, F.; Cope, L.; Hobbs, B.; Speed, T. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef] [PubMed]
- The Aroma Project. Available online: http://www.aroma-project.org (accessed on 3 June 2016).
- Ambroise, C.; McLachlan, G.J. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl. Acad. Sci. USA 2002, 99, 6562–6566. [Google Scholar] [CrossRef] [PubMed]
- Barla, A.; Mosci, S.; Rosasco, L.; Verri, A. A method for robust variable selection with significance assessment. In Proceedings of the ESANN 2008, Bruges, Belgium, 23–25 April 2008.
- PyXPlanner Documentation. Available online: http://slipguru.disi.unige.it/Software/PyXPlanner (accessed on 3 June 2016).
- Everitt, B. The Cambridge Dictionary of Statistics; Cambridge University Press: Cambridge, UK, 2006; p. 432. [Google Scholar]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B 2005, 67, 301–320. [Google Scholar]
- De Mol, C.; Mosci, S.; Traskine, M.; Verri, A. A Regularized Method for Selecting Nested Groups of Relevant Genes from Microarray Data. J. Comput. Biol. 2009, 16, 677–690. [Google Scholar] [CrossRef] [PubMed]
- De Mol, C.; De Vito, E.; Rosasco, L. Elastic Net Regularization in Learning Theory. J. Complex. 2009, 25, 201–230. [Google Scholar] [CrossRef]
- Fardin, P.; Barla, A.; Mosci, S.; Rosasco, L.; Verri, A.; Varesio, L. The l1-l2 regularization framework unmasks the hypoxia signature hidden in the transcriptome of a set of heterogeneous neuroblastoma cell lines. BMC Genom. 2009, 10, 474. [Google Scholar] [CrossRef] [PubMed]
- Fardin, P.; Barla, A.; Mosci, S.; Rosasco, L.; Verri, A.; Versteeg, R.; Caron, H.; Molenaar, J.; Ora, I.; Eva, A.; et al. A biology-driven approach identifies the hypoxia gene signature as a predictor of the outcome of neuroblastoma patients. Mol. Cancer 2010, 9, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squillario, M.; Barla, A. A computational procedure for functional characterization of potential marker genes from molecular data: Alzheimer’s as a case study. BMC Med. Genom. 2011, 4, 55. [Google Scholar] [CrossRef] [PubMed]
- Mascelli, S.; Barla, A.; Raso, A.; Mosci, S.; Nozza, P.; Biassoni, R.; Morana, G.; Huber, M.; Mircean, C.; Fasulo, D.; et al. Molecular fingerprinting reflects different histotypes and brain region in low grade gliomas. BMC Cancer 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- L1L2Signature Documentation. Available online: http://slipguru.disi.unige.it/Software/L1L2Signature (accessed on 3 June 2016).
- L1L2Py Documentation. Available online: http://slipguru.disi.unige.it/Software/L1L2Py (accessed on 3 June 2016).
- PPlus Documentation. Available online: http://slipguru.disi.unige.it/Software/PPlus (accessed on 3 June 2016).
- WebGESTALT Homepage. Available online: http://bioinfo.vanderbilt.edu/webgestalt/ (accessed on 3 June 2016).
- Zhang, B.; Kirov, S.; Snoddy, J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef] [PubMed]
- KDVS code repository. Available online: https://bitbucket.org/slipguru/kdvs (accessed on 3 June 2016).
- Zycinski, G.; Barla, A.; Verri, A. SVS: Data and knowledge integration in computational biology. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 6474–6478.
- Gene Ontology Consortium. Available online: http://geneontology.org/page/download-ontology (accessed on 3 June 2016).
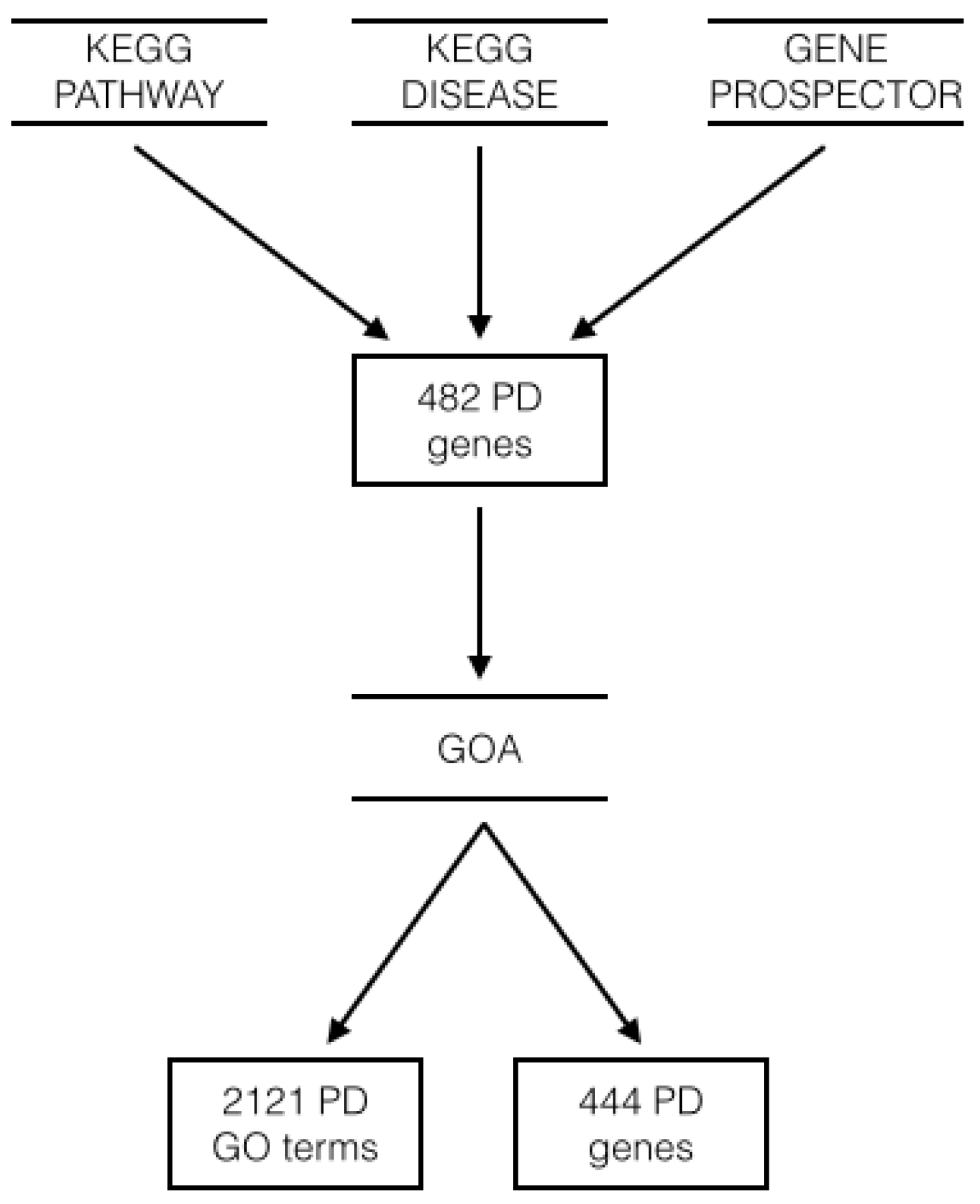
| Experiment | Test Error ± SD (%) | MCC |
|---|---|---|
| 23.1 ± 8.6 | 0.54 | |
| FiltKBest & LR | 22.0 ± 9.7 | 0.56 |
| LASSO & LR | 22.0 ± 8.2 | 0.56 |
| ENET & LR | 24.6 ± 7.1 | 0.51 |
| GO Terms | Genes | |||||
|---|---|---|---|---|---|---|
| Experiments | Precision (%) | Recall (%) | F-measure () | Precision (%) | Recall (%) | F-measure () |
| KDVS all domains | 44.0 | 12.7 | 197.4 | 7.5 | 25.5 | 115.5 |
| 71.4 | 0.2 | 4.8 | 10.4 | 1.1 | 20.4 | |
| FiltKBest & LR | 50.0 | 0.1 | 1.0 | 3.5 | 0.5 | 8.0 |
| LASSO & LR | 50.0 | 0.1 | 2.8 | 18.8 | 0.7 | 13.1 |
| ENET & LR | 62.5 | 0.2 | 4.8 | 16.7 | 0.9 | 17.1 |
| t-test | 50.0 | 0.1 | 1.0 | 2.5 | 0.2 | 4.2 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Squillario, M.; Barbieri, M.; Verri, A.; Barla, A. Enhancing Interpretability of Gene Signatures with Prior Biological Knowledge. Microarrays 2016, 5, 15. https://0-doi-org.brum.beds.ac.uk/10.3390/microarrays5020015
Squillario M, Barbieri M, Verri A, Barla A. Enhancing Interpretability of Gene Signatures with Prior Biological Knowledge. Microarrays. 2016; 5(2):15. https://0-doi-org.brum.beds.ac.uk/10.3390/microarrays5020015
Chicago/Turabian StyleSquillario, Margherita, Matteo Barbieri, Alessandro Verri, and Annalisa Barla. 2016. "Enhancing Interpretability of Gene Signatures with Prior Biological Knowledge" Microarrays 5, no. 2: 15. https://0-doi-org.brum.beds.ac.uk/10.3390/microarrays5020015