Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica napus L.) under Different Hormones and Abiotic Stress Conditions
Abstract
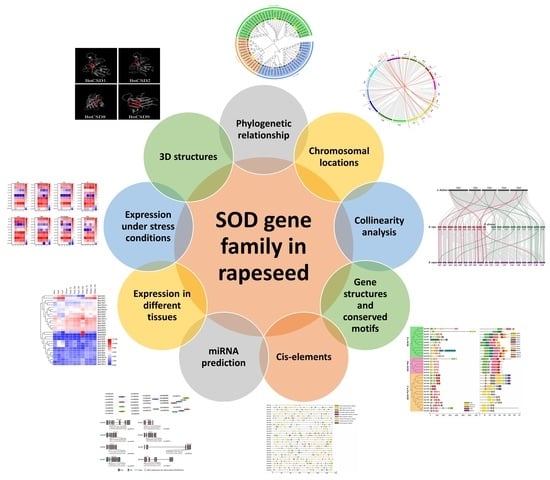
:1. Introduction
2. Materials and Methods
2.1. Identification and Characterization of SOD Genes in Rapeseed
2.2. Phylogenetic Tree and Synteny Analysis of BnSOD Proteins
2.3. Analysis of Cis-Acting Regulatory Elements in the BnSODs Promoters
2.4. Prediction of Putative miRNA Targeting BnSOD Genes and GO Annotation Analysis
2.5. Expression Profiling of BnSOD Genes in Different Tissues
2.6. Plant Material and Stress Conditions
2.7. RNA Extraction and qRT-PCR Analysis
2.8. Prediction of the 3D Structure of BnSOD Proteins
3. Results
3.1. Identification of SOD Gene Family in B. napus
3.2. Phylogenetic Relationships of SOD Genes
3.3. Chromosomal Locations and Synteny Analysis of SOD Genes
3.4. BnSOD Gene Structures and Conserved Motifs Investigation
3.5. Examination of Cis-Elements in Promoters of BnSOD Genes
3.6. Genome-Wide Analysis of miRNA Targeting BnSOD Genes
3.7. Functional Annotation Study of BnSOD Genes
3.8. Expression Analysis of BnSOD Genes in Several Developmental Tissues
3.9. Expression Profiles of BnSOD Genes under Hormones and Abiotic Stress Treatments
3.10. Prediction of the 3D Structures of BnSODs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Raza, A.; Ashraf, F.; Zou, X.; Zhang, X.; Tosif, H. Plant Adaptation and Tolerance to Environmental Stresses: Mechanisms and Perspectives. In Plant Ecophysiology and Adaptation under Climate Change: Mechanisms and Perspectives I; Springer: Singapore, 2020; pp. 117–145. [Google Scholar]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Hodgson, E.K.; Fridovich, I. Reversal of the superoxide dismutase reaction. Biochem. Biophys. Res. Commun. 1973, 54, 270–274. [Google Scholar] [CrossRef]
- Brawn, K.; Fridovich, I. Superoxide radical and superoxide dismutases: Threat and defense. In Autoxidation in Food and Biological Systems; Springer: Boston, MA, USA, 1980; pp. 429–446. [Google Scholar]
- Fink, R.C.; Scandalios, J.G. Molecular evolution and structure–function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 2002, 399, 19–36. [Google Scholar] [CrossRef]
- Abreu, I.A.; Cabelli, D.E. Superoxide dismutases—a review of the metal-associated mechanistic variations. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Neupane, K.; Shearer, J.; Palenik, B. Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ. Microbiol. 2008, 10, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Wang, W.; Yuan, R.; Den, F.; Shen, F. Superoxide dismutase and its research in plant stress-tolerance. Mol. Plant. Breed. 2015, 13, 2633–2646. [Google Scholar]
- Zeng, X.; Liu, Z.; Shi, P.; Xu, Y.; Sun, J.; Fang, Y.; Yang, G.; Wu, J.; Kong, D.; Sun, W. Cloning and expression analysis of copper and zinc superoxide dismutase (Cu/Zn-SOD) gene from Brassica campestris L. Acta Agron. Sin. 2014, 40, 636–643. [Google Scholar] [CrossRef]
- Song, J.; Zeng, L.; Chen, R.; Wang, Y.; Zhou, Y. In silico identification and expression analysis of superoxide dismutase (SOD) gene family in Medicago truncatula. 3 Biotech. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Perry, J.; Shin, D.; Getzoff, E.; Tainer, J. The structural biochemistry of the superoxide dismutases. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2010, 1804, 245–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alamri, S.; Hu, Y.; Mukherjee, S.; Aftab, T.; Fahad, S.; Raza, A.; Ahmad, M.; Siddiqui, M.H. Silicon-induced postponement of leaf senescence is accompanied by modulation of antioxidative defense and ion homeostasis in mustard (Brassica juncea) seedlings exposed to salinity and drought stress. Plant Physiol. Biochem. 2020, 157, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Chokshi, K.; Pancha, I.; Trivedi, K.; Maurya, R.; Ghosh, A.; Mishra, S. Physiological responses of the green microalga Acutodesmus dimorphus to temperature induced oxidative stress conditions. Physiol. Plant 2020, 170, 462–473. [Google Scholar] [CrossRef]
- Liu, T.; Ye, X.; Li, M.; Li, J.; Qi, H.; Hu, X. H2O2 and NO are involved in trehalose-regulated oxidative stress tolerance in cold-stressed tomato plants. Environ. Exp. Bot. 2020, 171, 103961. [Google Scholar] [CrossRef]
- Mosa, K.A.; El-Naggar, M.; Ramamoorthy, K.; Alawadhi, H.; Elnaggar, A.; Wartanian, S.; Ibrahim, E.; Hani, H. Copper nanoparticles induced genotoxicty, oxidative stress, and changes in Superoxide Dismutase (SOD) gene expression in cucumber (Cucumis sativus) plants. Front. Plant Sci. 2018, 9, 872. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Tian, W.; Huang, G.; Liu, B.; Wang, A.; Zhu, J.; Guo, X. The SikCuZnSOD3 gene improves abiotic stress resistance in transgenic cotton. Mol. Breed. 2021, 41, 1–17. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Omidi, M.; Naghavi, M.R.; Etminan, A.; Mehrabi, A.A.; Poczai, P. Wild Relatives of Wheat Respond Well to Water Deficit Stress: A Comparative Study of Antioxidant Enzyme Activities and Their Encoding Gene Expression. Agriculture 2020, 10, 415. [Google Scholar] [CrossRef]
- Lv, J.; Zhang, J.; Han, X.; Bai, L.; Xu, D.; Ding, S.; Ge, Y.; Li, C.; Li, J. Genome wide identification of superoxide dismutase (SOD) genes and their expression profiles under 1-methylcyclopropene (1-MCP) treatment during ripening of apple fruit. Sci. Hortic. 2020, 271, 109471. [Google Scholar] [CrossRef]
- Cui, C.; Wang, J.-J.; Zhao, J.-H.; Fang, Y.-Y.; He, X.-F.; Guo, H.-S.; Duan, C.-G. A Brassica miRNA Regulates Plant Growth and Immunity through Distinct Modes of Action. Mol. Plant 2020, 13, 231–245. [Google Scholar] [CrossRef]
- Khandal, H.; Parween, S.; Roy, R.; Meena, M.K.; Chattopadhyay, D. MicroRNA profiling provides insights into post-transcriptional regulation of gene expression in chickpea root apex under salinity and water deficiency. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Ding, L.; Xia, Y.; Wang, F.; Zhu, C. Emerging roles of microRNAs in plant heavy metal tolerance and homeostasis. J. Agric. Food Chem. 2020, 68, 1958–1965. [Google Scholar] [CrossRef]
- Ravichandran, S.; Ragupathy, R.; Edwards, T.; Domaratzki, M.; Cloutier, S. MicroRNA-guided regulation of heat stress response in wheat. BMC Genom. 2019, 20, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-Y.; Grabau, E. Bypassing miRNA-mediated gene regulation under drought stress: Alternative splicing affects CSD1 gene expression. Plant Mol. Biol. 2017, 95, 243–252. [Google Scholar] [CrossRef]
- Shi, G.-Q.; Fu, J.-Y.; Rong, L.-J.; Zhang, P.-Y.; Guo, C.-J.; Kai, X. TaMIR1119, a miRNA family member of wheat (Triticum aestivum), is essential in the regulation of plant drought tolerance. J. Integr. Agric. 2018, 17, 2369–2378. [Google Scholar] [CrossRef] [Green Version]
- Beauclair, L.; Yu, A.; Bouché, N. microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 2010, 62, 454–462. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Deng, F.; Yuan, R.; Shen, F. Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genom. 2017, 18, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Jin, X.; Zhu, Y.-X. Identification and analyses of miRNA genes in allotetraploid Gossypium hirsutum fiber cells based on the sequenced diploid G. raimondii genome. J. Genet. Genom. 2012, 39, 351–360. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Q.; Wang, K.; Pan, X.; Liu, F.; Guo, T.; Cobb, G.P.; Anderson, T.A. Identification of cotton microRNAs and their targets. Gene 2007, 397, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, L.; Yuan, D.; Lindsey, K.; Zhang, X. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J. Exp. Bot. 2013, 64, 1521–1536. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [Green Version]
- Raza, A. Eco-physiological and Biochemical Responses of Rapeseed (Brassica napus L.) to Abiotic Stresses: Consequences and Mitigation Strategies. J. Plant Growth Regul. 2020, 40, 1368–1388. [Google Scholar] [CrossRef]
- He, H.; Lei, Y.; Yi, Z.; Raza, A.; Zeng, L.; Yan, L.; Xiaoyu, D.; Yong, C.; Xiling, Z. Study on the mechanism of exogenous serotonin improving cold tolerance of rapeseed (Brassica napus L.) seedlings. Plant Growth Regul. 2021, 94, 161–170. [Google Scholar] [CrossRef]
- Mehmood, S.S.; Lu, G.; Luo, D.; Hussain, M.A.; Raza, A.; Zafar, Z.; Zhang, X.; Cheng, Y.; Zou, X.; Lv, Y. Integrated Analysis of Transcriptomics and Proteomics provides insights into the molecular regulation of cold response in Brassica napus. Environ. Exp. Bot. 2021, 187, 104480. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Hussain, M.A.; Wei, S.; He, H.; Zaman, Q.U.; Xuekun, Z.; Hasanuzzaman, M. Omics: The way forward to enhance abiotic stress tolerance in Brassica napus L. GM Crop. Food 2021, 12, 251–281. [Google Scholar] [CrossRef]
- Raza, A.; Su, W.; Gao, A.; Mehmood, S.S.; Hussain, M.A.; Nie, W.; Lv, Y.; Zou, X.; Zhang, X. Catalase (CAT) Gene Family in Rapeseed (Brassica napus L.): Genome-Wide Analysis, Identification, and Expression Pattern in Response to Multiple Hormones and Abiotic Stress Conditions. Inter. J. Mol. Sci. 2021, 22, 4281. [Google Scholar] [CrossRef]
- Song, J.M.; Liu, D.X.; Xie, W.Z.; Yang, Z.; Guo, L.; Liu, K.; Yang, Q.Y.; Chen, L.L. BnPIR: Brassica napus Pan-genome Information Resource for 1,689 accessions. Plant Biotechnol. J. 2021, 19, 412. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Beavis, W.; Berardini, T.Z.; Chen, G.; Dixon, D.; Doyle, A.; Garcia-Hernandez, M.; Huala, E.; Lander, G.; Montoya, M. The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003, 31, 224–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press Inc.: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [Green Version]
- Powell, S.; Forslund, K.; Szklarczyk, D.; Trachana, K.; Roth, A.; Huerta-Cepas, J.; Gabaldon, T.; Rattei, T.; Creevey, C.; Kuhn, M. eggNOG v4. 0: Nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014, 42, D231–D239. [Google Scholar] [CrossRef]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inform. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Wass, M.N.; Kelley, L.A.; Sternberg, M.J. 3DLigandSite: Predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010, 38, W469–W473. [Google Scholar] [CrossRef] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef]
- Xu, X.; Yang, Y.; Liu, C.; Sun, Y.; Zhang, T.; Hou, M.; Huang, S.; Yuan, H. The evolutionary history of the sucrose synthase gene family in higher plants. BMC Plant Biol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Song, J.-M.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Chen, J.; Li, R.; Shang, S.; Tang, X. Genome-wide analysis of the superoxide dismutase (SOD) gene family in Zostera marina and expression profile analysis under temperature stress. PeerJ 2020, 8, e9063. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Chen, Y.; Wang, S.; Fang, Y.; Zhang, X.; Wu, Y.; Xue, D. Genome-wide identification of the SOD gene family and expression analysis under drought and salt stress in barley. Plant Growth Regul. 2021, 94, 49–60. [Google Scholar] [CrossRef]
- Filiz, E.; Tombuloğlu, H. Genome-wide distribution of superoxide dismutase (SOD) gene families in Sorghum bicolor. Turk. J. Biol. 2015, 39, 49–59. [Google Scholar] [CrossRef]
- Feng, K.; Yu, J.; Cheng, Y.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Li, Z.; Yao, Z.; Yang, Y. The SOD gene family in tomato: Identification, phylogenetic relationships, and expression patterns. Front. Plant Sci. 2016, 7, 1279. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Hao, C.; Cheng, Z.-M.; Zhong, Y. Genome-wide identification, characterization, and expression analysis of the grapevine superoxide dismutase (SOD) family. Inter. J. Genom. 2019, 2019, 7350414. [Google Scholar] [CrossRef]
- Feng, X.; Lai, Z.; Lin, Y.; Lai, G.; Lian, C. Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genom. 2015, 16, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Yang, L.; He, Y.; Zhang, H.; Li, W.; Chen, H.; Ma, D.; Yin, J. Genome-wide identification and transcriptional expression analysis of superoxide dismutase (SOD) family in wheat (Triticum aestivum). PeerJ 2019, 7, e8062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genom. 2019, 20, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Xia, M.; Chen, J.; Deng, F.; Yuan, R.; Zhang, X.; Shen, F. Data set for phylogenetic tree and RAMPAGE Ramachandran plot analysis of SODs in Gossypium raimondii and G. arboreum. Data Brief. 2016, 9, 345–348. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Nati. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [Green Version]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Nakamura, Y.; Watanabe-Takahashi, A.; Inoue, E.; Yamaya, T.; Takahashi, H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005, 42, 305–314. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.; Wu, H.; Jiang, L.; Liu, S. Genome-wide identification and transcriptional expression analysis of cucumber superoxide dismutase (SOD) family in response to various abiotic stresses. Inter. J. Genom. 2017, 2017, 7243973. [Google Scholar] [CrossRef]
- Chen, L.; Chen, L.; Zhang, X.; Liu, T.; Niu, S.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T. Identification of miRNAs that regulate silique development in Brassica napus. Plant Sci. 2018, 269, 106–117. [Google Scholar] [CrossRef]
- Buhtz, A.; Springer, F.; Chappell, L.; Baulcombe, D.C.; Kehr, J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 2008, 53, 739–749. [Google Scholar] [CrossRef]
- Fu, Y.; Mason, A.S.; Zhang, Y.; Lin, B.; Xiao, M.; Fu, D.; Yu, H. MicroRNA-mRNA expression profiles and their potential role in cadmium stress response in Brassica napus. BMC Plant Biol. 2019, 19, 1–20. [Google Scholar] [CrossRef]
- Körbes, A.P.; Machado, R.D.; Guzman, F.; Almerao, M.P.; de Oliveira, L.F.V.; Loss-Morais, G.; Turchetto-Zolet, A.C.; Cagliari, A.; dos Santos Maraschin, F.; Margis-Pinheiro, M. Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PLoS ONE 2012, 7, e50663. [Google Scholar] [CrossRef]
- Shen, E.; Zou, J.; Hubertus Behrens, F.; Chen, L.; Ye, C.; Dai, S.; Li, R.; Ni, M.; Jiang, X.; Qiu, J. Identification, evolution, and expression partitioning of miRNAs in allopolyploid Brassica napus. J. Exp. Bot. 2015, 66, 7241–7253. [Google Scholar] [CrossRef] [Green Version]
- Casati, P. Analysis of UV-B regulated miRNAs and their targets in maize leaves. Plant Signal. Behav. 2013, 8, e26758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Yu, X.; Lei, N.; Cheng, Z.; Zhao, P.; He, Y.; Wang, W.; Peng, M. Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Sci. Rep. 2017, 7, 45981. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Qiao, Y.; Zhang, J.; Shi, W.; Zhang, J. Genome wide identification of microRNAs involved in fatty acid and lipid metabolism of Brassica napus by small RNA and degradome sequencing. Gene 2017, 619, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jian, H.; Wang, T.; Wei, L.; Li, J.; Li, C.; Liu, L. Identification of microRNAs actively involved in fatty acid biosynthesis in developing Brassica napus seeds using high-throughput sequencing. Front. Plant Sci. 2016, 7, 1570. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.-H.; Helliwell, C.A. Regulation of flowering time and floral patterning by miR172. J. Exp. Bot. 2011, 62, 487–495. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.-W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.B.; Gao, S.; Sun, D.; Li, H.; Shu, X.X.; Yang, Z.M. miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol. 2013, 13, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.B.; Gao, S.; Wang, Y.; Li, B.W.; Zhang, Y.L.; Yang, Z.M. miR394 and its target gene LCR are involved in cold stress response in Arabidopsis. Plant Gene 2016, 5, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Xia, M.; Chen, J.; Deng, F.; Yuan, R.; Zhang, X.; Shen, F. Genome-wide analysis of superoxide dismutase gene family in Gossypium raimondii and G. arboreum. Plant Gene 2016, 6, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Dehury, B.; Sarma, K.; Sarmah, R.; Sahu, J.; Sahoo, S.; Sahu, M.; Sen, P.; Modi, M.K.; Sharma, G.D.; Choudhury, M.D. In silico analyses of superoxide dismutases (SODs) of rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 2013, 22, 150–156. [Google Scholar] [CrossRef]
- Fisher, C.L.; Cabelli, D.E.; Tainer, J.A.; Hallewell, R.A.; Getzoff, E.D. The role of arginine 143 in the electrostatics and mechanism of Cu, Zn superoxide dismutase: Computational and experimental evaluation by mutational analysis. Proteins Struct. Funct. Bioinform. 1994, 19, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Ramana Gopavajhula, V.; Viswanatha Chaitanya, K.; Akbar Ali Khan, P.; Shaik, J.P.; Narasimha Reddy, P.; Alanazi, M. Modeling and analysis of soybean (Glycine max. L) Cu/Zn, Mn and Fe superoxide dismutases. Genet. Mol. Biol. 2013, 36, 225–236. [Google Scholar] [CrossRef]









| Gene ID | Gene Name | Genomic Position (bp) | Gene Length (bp) | CDS Length (bp) | Exons | Protein Length (Amino Acids) | Molecular Weight (kDa) | Isoelectric Point (pI) | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| BnaA04T0182200ZS | BnCSD1 | A04-18599782:18600942 (+) | 1160 | 639 | 6 | 212 | 22.15 | 7.16 | Chloroplast |
| BnaA06T0053200ZS | BnCSD2 | A06-3364923:3368393 (+) | 3470 | 1173 | 9 | 390 | 42.34 | 8.71 | Cytoskeleton |
| BnaA08T0278700ZS | BnCSD3 | A08-26336887:26338407 (+) | 1520 | 954 | 6 | 317 | 33.76 | 5.19 | Chloroplast |
| BnaA09T0129500ZS | BnCSD4 | A09-7757584:7758410 (−) | 826 | 660 | 3 | 219 | 23.60 | 5.95 | Chloroplast |
| BnaA09T0647100ZS | BnCSD5 | A09-61965958:61967448 (+) | 1490 | 954 | 6 | 317 | 33.45 | 5.26 | Chloroplast |
| BnaA09T0664700ZS | BnCSD6 | A09-62938095:62939938 (−) | 1843 | 459 | 8 | 152 | 15.17 | 5.64 | Cytoskeleton |
| BnaA10T0190600ZS | BnCSD7 | A01-21329437:21330797 (−) | 1360 | 465 | 6 | 154 | 15.94 | 7.12 | Cytoskeleton |
| BnaC04T0482300ZS | BnCSD8 | C04-60854831:60856275 (+) | 1444 | 627 | 6 | 208 | 21.60 | 7.84 | Chloroplast |
| BnaC05T0066000ZS | BnCSD9 | C05-3767908:3769181 (+) | 1273 | 441 | 7 | 146 | 14.55 | 5.44 | Cytoskeleton |
| BnaC08T0217500ZS | BnCSD10 | C08-32001876:32003366 (−) | 1490 | 951 | 5 | 316 | 33.71 | 6.91 | Chloroplast |
| BnaC08T0505400ZS | BnCSD11 | C08-51947641:51949188 (+) | 1547 | 957 | 6 | 318 | 33.58 | 4.97 | Chloroplast |
| BnaC08T0529200ZS | BnCSD12 | C08-53323394:53325251 (−) | 1857 | 672 | 7 | 223 | 23.42 | 6.66 | Cytoskeleton |
| BnaC09T0137500ZS | BnCSD13 | C09-10172656:10173492 (−) | 836 | 660 | 3 | 219 | 23.72 | 6.83 | Chloroplast |
| BnaC09T0484500ZS | BnCSD14 | C09-59599637:59601046 (−) | 1409 | 510 | 6 | 169 | 17.63 | 6.82 | Cytoskeleton |
| BnaA01T0146300ZS | BnFSD1 | A01-8654405:8657035 (−) | 2630 | 561 | 6 | 186 | 21.12 | 6.06 | Cytoskeleton |
| BnaA03T0141400ZS | BnFSD2 | A03-7188761:7190553 (+) | 1792 | 906 | 8 | 301 | 34.51 | 4.97 | Chloroplast |
| BnaA06T0320600ZS.1 | BnFSD3 | A06-40740605:40744473 (+) | 3868 | 879 | 9 | 292 | 33.60 | 7.58 | Chloroplast |
| BnaA09T0072700ZS | BnFSD4 | A09-4315462:4316968 (−) | 1506 | 792 | 8 | 263 | 30.16 | 7.76 | Chloroplast |
| BnaA10T0083700ZS | BnFSD5 | A10-13303579:13310477 (−) | 6898 | 738 | 6 | 245 | 28.02 | 9.44 | Mitochondrion |
| BnaC01T0186100ZS | BnFSD6 | C01-13777631:13779146 (−) | 1515 | 618 | 5 | 205 | 22.99 | 6.3 | Cytoskeleton |
| BnaC03T0164000ZS | BnFSD7 | C03-9109698:9111720 (+) | 2022 | 903 | 9 | 300 | 34.39 | 4.88 | Chloroplast |
| BnaC07T0373300ZS | BnFSD8 | C07-49894566:49896148 (−) | 1582 | 780 | 6 | 259 | 29.58 | 8.58 | Chloroplast |
| BnaC07T0462000ZS | BnFSD9 | C07-55868729:55870093 (−) | 1364 | 648 | 5 | 215 | 24.17 | 6.65 | Endoplasmic reticulum |
| BnaC09T0062700ZS | BnFSD10 | C09-4091989:4094054 (−) | 2065 | 783 | 8 | 260 | 29.72 | 6.66 | Chloroplast |
| BnaC09T0329400ZS | BnFSD11 | C09-41111988:41116227 (−) | 4239 | 1077 | 7 | 358 | 40.40 | 8.8 | Chloroplast |
| BnaA01T0376200ZS | BnMSD1 | A01-33774266:33775498 (−) | 1232 | 699 | 6 | 232 | 25.50 | 8.38 | Mitochondrion |
| BnaA05T0446100ZS | BnMSD2 | A05-41549634:41550932 (−) | 1298 | 696 | 6 | 231 | 25.41 | 8.47 | Mitochondrion |
| BnaA09T0519400ZS | BnMSD3 | A09-55258451:55262461 (−) | 4010 | 966 | 9 | 321 | 35.60 | 9.56 | Cytoskeleton |
| BnaC01T0471300ZS | BnMSD4 | C01-53548352:53549620 (−) | 1268 | 693 | 6 | 230 | 25.36 | 8.94 | Mitochondrion |
| BnaC05T0492700ZS | BnMSD5 | C05-53531802:53533097 (−) | 1295 | 696 | 6 | 231 | 25.43 | 7.83 | Mitochondrion |
| BnaC08T0362400ZS | BnMSD6 | C08-43431847:43433140 (−) | 1293 | 729 | 6 | 242 | 27.11 | 6.14 | Endoplasmic reticulum |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, W.; Raza, A.; Gao, A.; Jia, Z.; Zhang, Y.; Hussain, M.A.; Mehmood, S.S.; Cheng, Y.; Lv, Y.; Zou, X. Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica napus L.) under Different Hormones and Abiotic Stress Conditions. Antioxidants 2021, 10, 1182. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10081182
Su W, Raza A, Gao A, Jia Z, Zhang Y, Hussain MA, Mehmood SS, Cheng Y, Lv Y, Zou X. Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica napus L.) under Different Hormones and Abiotic Stress Conditions. Antioxidants. 2021; 10(8):1182. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10081182
Chicago/Turabian StyleSu, Wei, Ali Raza, Ang Gao, Ziqi Jia, Yi Zhang, Muhammad Azhar Hussain, Sundas Saher Mehmood, Yong Cheng, Yan Lv, and Xiling Zou. 2021. "Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica napus L.) under Different Hormones and Abiotic Stress Conditions" Antioxidants 10, no. 8: 1182. https://0-doi-org.brum.beds.ac.uk/10.3390/antiox10081182