Alpha and Beta-diversity of Microbial Communities Associated to Plant Disease Suppressive Functions of On-farm Green Composts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Compost Collection
2.2. Compost Suppressive Assay
2.3. Assessment of Compost Phytotoxicity
2.4. Microbiologial Characterization of Compost
2.4.1. Population Counting
2.4.2. Enzymatic and Metabolic Analysis
2.5. T-RFLP Analysis
2.6. Biodiversity Assessment and Statistical Analysis
3. Results
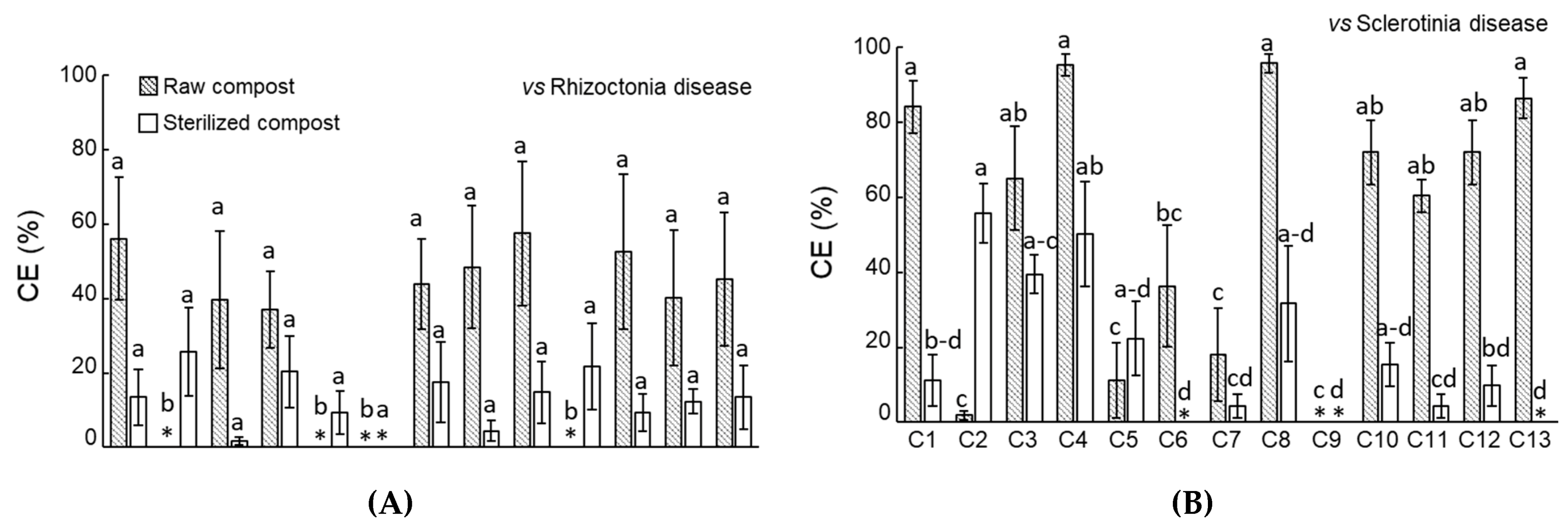
3.1. Compost Disease Suppressiveness
3.2. Compost Microbiological Features
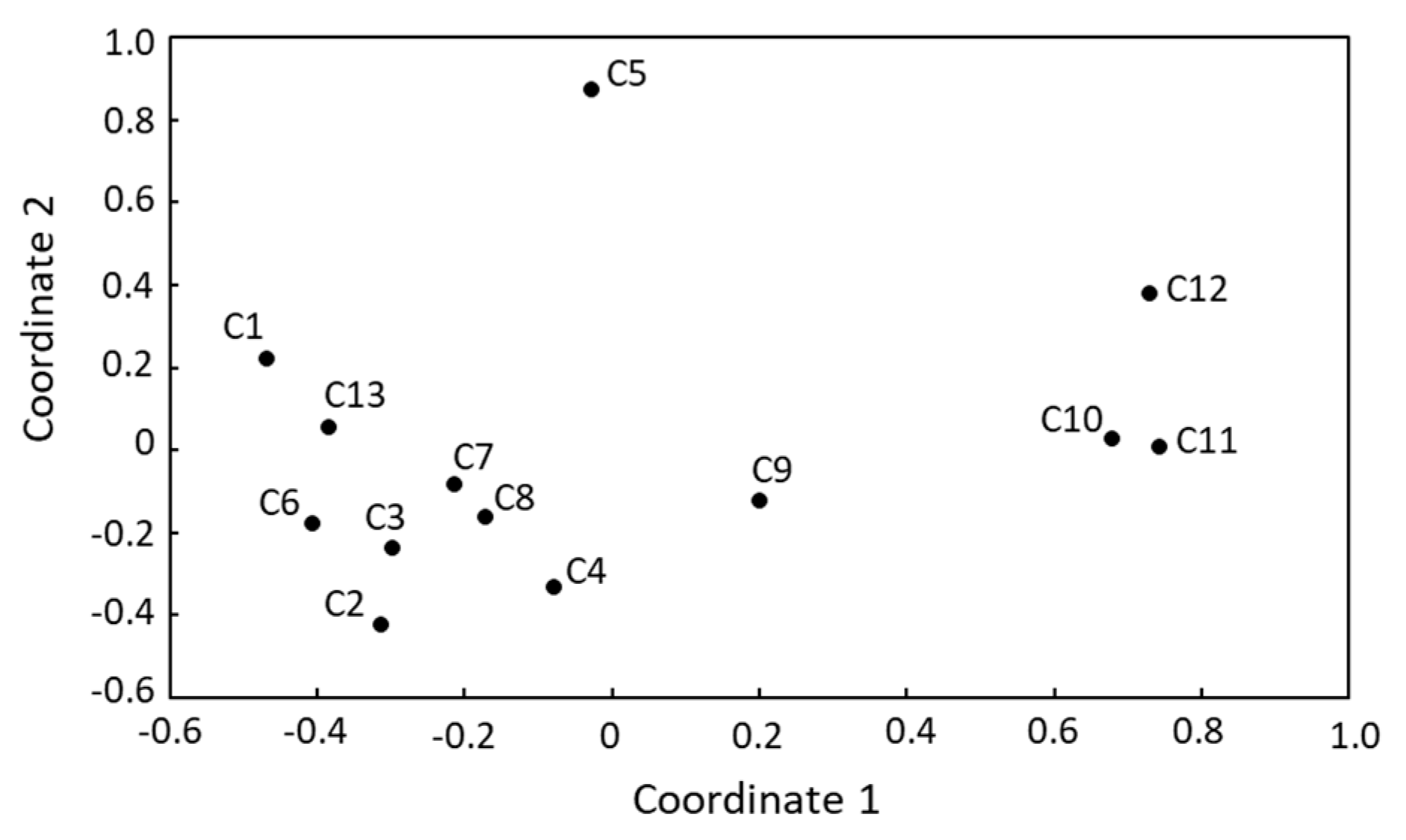
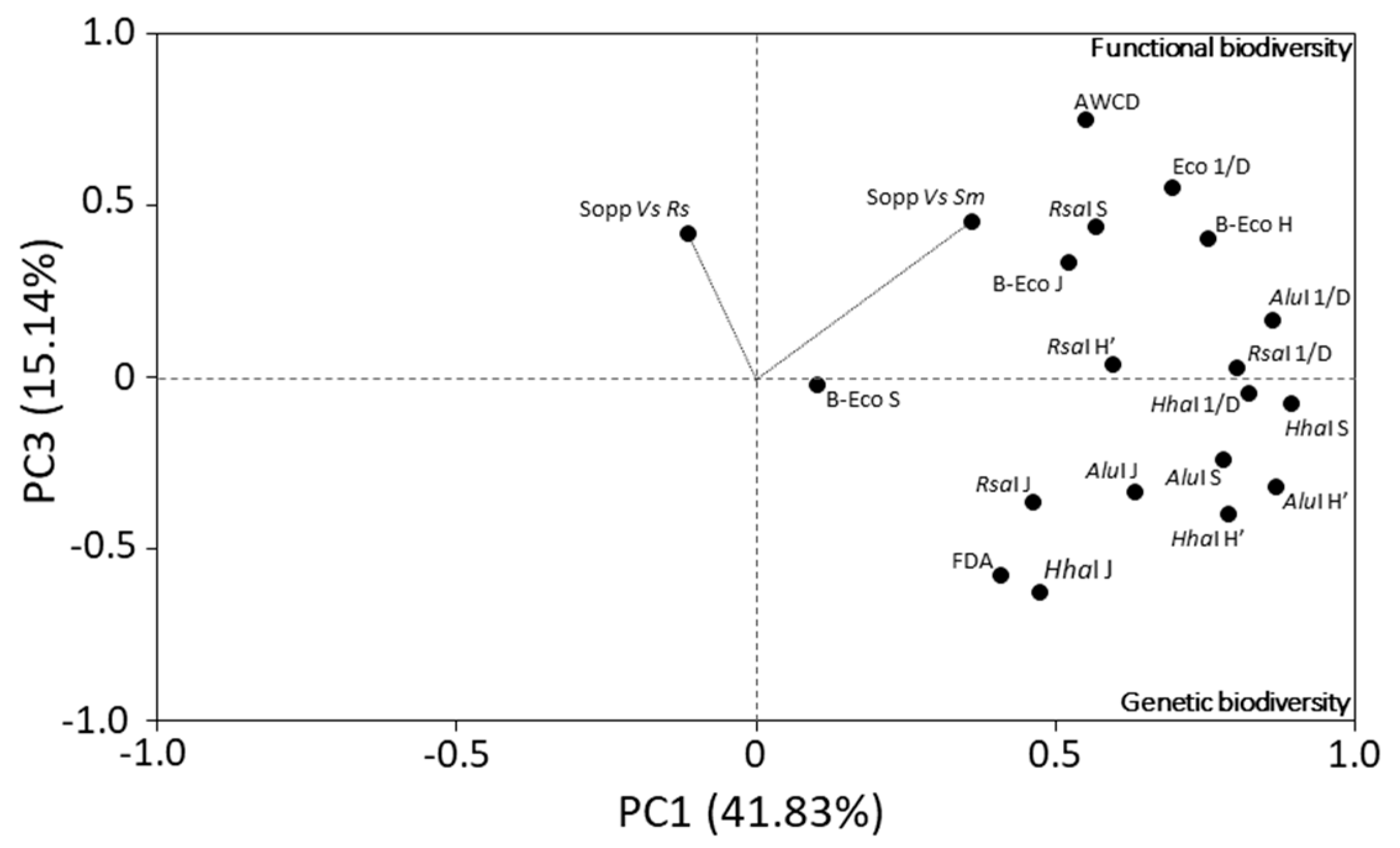
3.3. Microbial Biodiversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- Bonilla, N.; Gutiérrez-Barranquero, J.A.; Vicente, A.D.; Cazorla, F.M. Enhancing soil quality and plant health through suppressive organic amendments. Diversity 2012, 4, 475–491. [Google Scholar] [CrossRef]
- Pane, C.; Spaccini, R.; Piccolo, A.; Scala, F.; Bonanomi, G. Compost amendments enhance peat suppressiveness to Pythium ultimum, Rhizoctonia solani and Sclerotinia minor. Biol. Control 2011, 56, 115–124. [Google Scholar] [CrossRef]
- Hadar, Y.; Papadopoulou, K.K. Suppressive composts: Microbial ecology links between abiotic environments and healthy plants. Annu. Rev. Phytopathol. 2012, 50, 133–153. [Google Scholar] [CrossRef] [PubMed]
- Stavi, I.; Bel, G.; Zaady, E. Soil functions and ecosystem services in conventional, conservation, and integrated agricultural systems. A review. Agron. Sustain. Dev. 2016, 36, 32. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, K.; Hartemink, A.E. Linking soils to ecosystem services—A global review. Geoderma 2016, 262, 101–111. [Google Scholar] [CrossRef]
- Pal, K.K.; McSpadden Gardener, B. Biological Control of Plant Pathogens. In The Plant Health Instructor; APSnet: Saint Paul, MN, USA, 2006; pp. 1–25. [Google Scholar]
- Scotti, R.; Pane, C.; Spaccini, R.; Palese, A.M.; Piccolo, A.; Celano, G.; Zaccardelli, M. On-farm compost: A useful tool to improve soil quality under intensive farming systems. Appl. Soil Ecol. 2016, 107, 13–23. [Google Scholar] [CrossRef]
- Pane, C.; Piccolo, A.; Spaccini, R.; Celano, G.; Villecco, D.; Zaccardelli, M. Agricultural waste-based composts exhibiting suppressivity to diseases caused by the phytopathogenic soil-borne fungi Rhizoctonia solani and Sclerotinia minor. Appl. Soil Ecol. 2013, 65, 43–51. [Google Scholar] [CrossRef]
- Pane, C.; Spaccini, R.; Piccolo, A.; Celano, G.; Zaccardelli, M. Disease suppressiveness of agricultural greenwaste composts as related to chemical and bio-based properties shaped by different on-farm composting methods. Biol. Control 2019, 137, 104026. [Google Scholar] [CrossRef]
- Scotti, R.; Bertora, C.; Pastore, V.; Antonucci, M.; Pane, C.; Gaudino, S.; Persiani, A.; Sorrentino, R.; Di Meo, V.; Grignani, C.; et al. Life Carbonfarm project: Technologies to stabilize soil organic carbon and farm productivity, promote waste value and climate change mitigation. In Proceedings of the Global Symposium on Soil Organic Carbon, Rome, Italy, 21–23 March 2017. [Google Scholar]
- Nakasaki, K.; Hirai, H.; Mimoto, H.; Quyen, T.N.M.; Koyama, M.; Takeda, K. Succession of microbial community during vigorous organic matter degradation in the primary fermentation stage of food waste composting. Sci. Total Environ. 2019, 671, 1237–1244. [Google Scholar] [CrossRef]
- Shin, T.S.; Yu, N.H.; Lee, J.; Choi, G.J.; Kim, J.C.; Shin, C.S. Development of a biofungicide using a mycoparasitic fungus Simplicillium lamellicola BCP and its control efficacy against gray mold diseases of tomato and ginseng. Plant Pathol. J. 2017, 33, 337–344. [Google Scholar] [CrossRef]
- Bartelt-Ryser, J.; Joshi, J.; Schmid, B.; Brandl, H.; Balser, T. Soil feedbacks of plant diversity on soil microbial communities and subsequent plant growth. Perspect. Plant Ecol. Evol. Syst. 2005, 7, 27–49. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tipayno, S.; Kim, C.G.; Sa, T. T-RFLP analysis of structural changes in soil bacterial communities in response to metal and metalloid contamination and initial phytoremediation. Appl. Soil Ecol. 2012, 61, 137–146. [Google Scholar] [CrossRef]
- Babendreier, D.; Joller, D.; Romeis, J.; Bigler, F.; Widmer, F. Bacterial community structures in honeybee intestines and their response to two insecticidal proteins. FEMS Microbiol. Ecol. 2007, 59, 600–610. [Google Scholar] [CrossRef]
- Blaud, A.; Diouf, F.; Herrmann, A.M.; Lerch, T.Z. Analysing the effect of soil organic matter on bacterial communities using T-RFLP fingerprinting: Different methods, different stories? Biol. Fertil. Soils 2015, 51, 959. [Google Scholar] [CrossRef] [Green Version]
- Zucconi, F.; Pera, A.; Forte, M.; de Bertoldi, M. Evaluating toxicity of immature compost. BioCycle 1981, 22, 54–57. [Google Scholar]
- Hadar, Y. Suppressive compost: When plant pathology met microbial ecology. Phytoparasitica 2011, 39, 311. [Google Scholar] [CrossRef] [Green Version]
- Malandraki, I.; Tjamos, S.E.; Pantelides, I.S.; Paplomatas, E.J. Thermal inactivation of compost suppressiveness implicates possible biological factors in disease management. Biol. Control 2008, 44, 180–187. [Google Scholar] [CrossRef]
- Flores-Rentería, D.; Rincón, A.; Valladaresa, F.; Curiel Yuste, J. Agricultural matrix affects differently the alpha and beta structural and functional diversity of soil microbial communities in a fragmented Mediterranean holm oak forest. Soil Biol. Biochem. 2016, 92, 79–90. [Google Scholar] [CrossRef]
- Robledo-Mahón, T.; Aranda, E.; Pesciaroli, C.; Rodríguez-Calvo, A.; Silva-Castro, G.A.; González-López, J. Effect of semi-permeable cover system on the bacterial diversity during sewage sludge composting. J. Environ. Manag. 2018, 215, 57–67. [Google Scholar] [CrossRef]
- Cao, G.; Song, T.; Shen, Y.; Jin, Q.; Feng, W.; Fan, L.; Cai, W. Diversity of bacterial and fungal communities in wheat straw compost for Agaricus bisporus cultivation. HortScience 2019, 54, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Esan, E.O.; Abbey, L.; Yurgel, S. Exploring the long-term effect of plastic on compost microbiome. PLoS ONE 2019, 14, e0214376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xun, W.; Li, W.; Xiong, W.; Ren, Y.; Liu, Y.; Miao, Y.; Xu, Z.; Zhang, N.; Shen, Q.; Zhang, R. Diversity-triggered deterministic bacterial assembly constrains community functions. Nat. Commun. 2019, 10, 3833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutgers, M.; Wouterse, M.; Drost, S.M.; Breure, A.M.; Mulder, C.; Stone, D.; Creamer, R.E.; Winding, A.; Bloem, J. Monitoring soil bacteria with community-level physiological profiles using Biolog™ ECO-plates in the Netherlands and Europe. Appl. Soil Ecol. 2016, 97, 23–35. [Google Scholar] [CrossRef]
- Zhou, D.; Jing, T.; Chen, Y.; Wang, F.; Qi, D.; Feng, R.; Xie, J.; Li, H. Deciphering microbial diversity associated with Fusarium wilt-diseased and disease free banana rhizosphere soil. BMC Microbiol. 2019, 19, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, C.M.; Pudake, R.N.; Srivastava, R.; Palni, U.; Sharma, A.K. Development of PCR-based molecular marker for screening of disease-suppressive composts against Fusarium wilt of tomato (Solanum lycopersicum L.). 3 Biotech 2018, 8, 306. [Google Scholar] [CrossRef] [PubMed]
- Weller, D.M.; Raaijmakers, J.M.; McSpadden Gardener, B.B.; Thomashow, L.S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 2002, 40, 309–348. [Google Scholar] [CrossRef] [Green Version]
- Zaccardelli, M.; De Nicola, F.; Villecco, D.; Scotti, R. The development and suppressive activity of soil microbial communities under compost amendment. J. Soil Sci. Plant Nutr. 2013, 13, 730–742. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Tian, Y.; Gao, Y.; Li, J. Microbial diversity in compost is critical in suppressing plant fungal pathogen survival and enhancing cucumber seedling growth. Compost Sci. Util. 2018, 26, 189–200. [Google Scholar] [CrossRef]
- Blaya, J.; Marhuenda, F.C.; Pascual, J.A.; Ros, M. Microbiota characterization of compost using omics approaches opens new perspectives for Phytophthora root rot control. PLoS ONE 2016, 11, e0158048. [Google Scholar] [CrossRef] [Green Version]
- Blaya, J.; Lloret, E.; Ros, M.; Pascual, J.A. Identification of predictor parameters to determine agro-industrial compost suppressiveness against Fusarium oxysporum and Phytophthora capsici diseases in muskmelon and pepper seedlings. J. Sci. Food Agric. 2014, 95, 1482–1490. [Google Scholar] [CrossRef] [PubMed]
- Vida, C.; Bonilla, N.; de Vicente, A.; Cazorla, F.M. Microbial profiling of a suppressiveness-induced agricultural soil amended with composted almond shells. Front Microbiol. 2016, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- De Corato, U.; Salimbeni, R.; De Pretis, A.; Patruno, L.; Avella, N.; Lacolla, G.; Cucci, G. Microbiota from ‘next-generation green compost’ improves suppressiveness of composted Municipal-Solid-Waste to soil-borne plant pathogens. Biol. Control 2018, 124, 1–17. [Google Scholar] [CrossRef]
- De Corato, U.; Patruno, L.; Avella, N.; Lacolla, G.; Cucci, G. Composts from green sources show an increased suppressiveness to soilborne plant pathogenic fungi: Relationships between physiochemical properties, disease suppression, and the microbiome. Crop Prot. 2019, 124, 104870. [Google Scholar] [CrossRef]
- Pane, C.; Zaccardelli, M. Principles of compost-based plant diseases control and innovative new developments. In Composting for Sustainable Agriculture; Maheshwari, D.K., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2014; Volume 3, pp. 151–171. [Google Scholar]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pane, C.; Sorrentino, R.; Scotti, R.; Molisso, M.; Di Matteo, A.; Celano, G.; Zaccardelli, M. Alpha and Beta-diversity of Microbial Communities Associated to Plant Disease Suppressive Functions of On-farm Green Composts. Agriculture 2020, 10, 113. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture10040113
Pane C, Sorrentino R, Scotti R, Molisso M, Di Matteo A, Celano G, Zaccardelli M. Alpha and Beta-diversity of Microbial Communities Associated to Plant Disease Suppressive Functions of On-farm Green Composts. Agriculture. 2020; 10(4):113. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture10040113
Chicago/Turabian StylePane, Catello, Roberto Sorrentino, Riccardo Scotti, Marcella Molisso, Antonio Di Matteo, Giuseppe Celano, and Massimo Zaccardelli. 2020. "Alpha and Beta-diversity of Microbial Communities Associated to Plant Disease Suppressive Functions of On-farm Green Composts" Agriculture 10, no. 4: 113. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture10040113





