Establishment of a Bivector Genetic Transformation System in Recalcitrant Maize Inbred Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Bivector Construction
2.3. Microprojectile Bombardment Technology
2.4. Agrobacterium-Mediated Transformation
2.5. Culture Media and Conditions for the Plant Genetic Transformation System
2.6. Genomic DNA Extraction and PCR Assay
2.7. Southern Blot Analyses
2.8. RNA Extraction and RT-qPCR Analysis
3. Results
3.1. Bivector Construction for Genetic Transformation of Recalcitrant Maize Inbred Lines
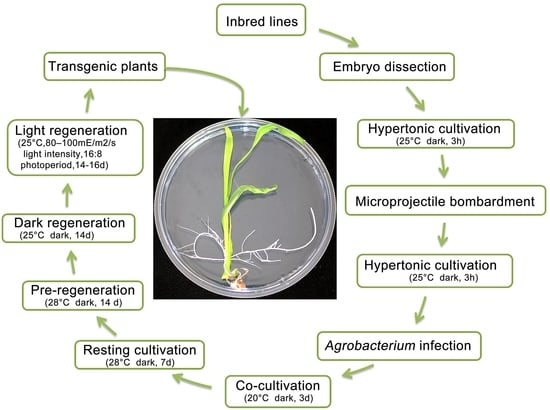
3.2. Culture Process of Genetic Transformation in Recalcitrant Maize Inbred Lines
3.3. Transformation Frequencies of Maize Recalcitrant Inbred Lines
3.4. Progeny Analysis of Transgenic Zheng58 Plants
3.5. Progeny Segregation Analysis of Transgenic Zheng58 Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yao, D.; Qi, W.; Li, X.; Yang, Q.; Yan, S.; Ling, H.; Wang, D.; Wang, G.; Song, R. Maize opaque10 encodes a cereal-specific protein that is essential for the proper distribution of zeins in en-dosperm protein bodies. PLoS Genet. 2016, 12, e1006270. [Google Scholar] [CrossRef]
- Ishida, Y.; Hiei, Y.; Komari, T. Tissue culture protocols for gene transfer and editing in maize (Zea mays L.). Plant Biotechnol. 2020, 37, 121–128. [Google Scholar] [CrossRef]
- Yadava, P.; Abhishek, A.; Singh, R.; Singh, I.; Kaul, T.; Pattanayak, A.; Agrawal, P.K. Advances in maize transformation technologies and development of transgenic maize. Front. Plant Sci. 2017, 7, 1949. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, C.L.; Green, C.E. Establishment and maintenance of friable, embryogenic maize callus and the involvement of L-proline. Planta 1985, 164, 207–214. [Google Scholar] [CrossRef]
- Armstrong, C.L.; Green, C.E.; Phillips, R. Development and availability of germplasm with high Type II culture formation response. Maize Genet. Coop. News Lett. 1991, 65, 92–93. [Google Scholar]
- Vega, J.; Yu, W.; Kennon, A.R.; Chen, X.; Zhang, Z.J. Improvement of agrobacterium-mediated transformation in HI-II maize (Zea mays) using standard binary vectors. Plant Cell Rep. 2007, 27, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Saito, H.; Ohta, S.; Hiei, Y.; Komari, T.; Kumashiro, T. High efficiency transformation of maize (Zea mays L.) mediated by agrobacterium tumefaciens. Nat. Biotechnol. 1996, 14, 745–750. [Google Scholar] [CrossRef]
- Abhishek, A.; Kumari, R.; Karjagi, C.G.; Kumar, P.; Kumar, B.; Dass, S.; Kumar, R.S.; Ramteke, P.W. Tissue culture independent agrobacterium tumefaciens mediated in planta transformation method for tropical maize (Zea mays L.). Proc. Natl. Acad. Sci. India Sect. B Boil. Sci. 2016, 86, 375–384. [Google Scholar] [CrossRef]
- Jones, T.; Lowe, K.; Hoerster, G.; Anand, A.; Wu, E.; Wang, N.; Arling, M.; Lenderts, B. Maize transformation using the morphogenic genes baby boom and wuschel2. In Transgenic Plants: Methods and Protocols; Kumar, S., Barone, P., Smith, M., Eds.; Springer: New York, NY, USA, 2019; pp. 81–93. [Google Scholar]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; van Lammeren, A.A.M.; Miki, B.L.A.; et al. Ectopic expression of baby boom triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, C.; Liu, Z.; Heidmann, I.; Supena, E.D.J.; Fukuoka, H.; Joosen, R.; Lambalk, J.; Angenent, G.; Scorza, R.; Custers, J.B.M.; et al. Heterologous expression of the baby boom AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta 2006, 225, 341–351. [Google Scholar] [CrossRef]
- Heidmann, I.; De Lange, B.; Lambalk, J.; Angenent, G.C.; Boutilier, K. Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep. 2011, 30, 1107–1115. [Google Scholar] [CrossRef] [Green Version]
- Florez, S.L.; Erwin, R.L.; Maximova, S.N.; Guiltinan, M.J.; Curtis, W.R. Enhanced somatic embryogenesis in Theobroma cacao using the homologous baby boom transcription factor. BMC Plant Biol. 2015, 15, 121. [Google Scholar] [CrossRef] [Green Version]
- Laux, T.; Mayer, K.; Berger, J.; Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef]
- Rashid, S.Z.; Yamaji, N.; Kyo, M. Shoot formation from root tip region: A developmental alteration by WUS in transgenic tobacco. Plant Cell Rep. 2007, 26, 1449–1455. [Google Scholar] [CrossRef]
- Arroyo-Herrera, A.; Gonzalez, A.K.; Moo, R.C.; Quiroz-Figueroa, F.R.; Loyola-Vargas, V.M.; Rodriguez-Zapata, L.; D′hondt, C.B.; Suárez-Solís, V.M.; Castaño, E. Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tissue Organ. Cult. 2008, 94, 171–180. [Google Scholar] [CrossRef]
- Bouchabké-Coussa, O.; Obellianne, M.; Linderme, D.; Montes, E.; Maia-Grondard, A.; Vilaine, F.; Pannetier, C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013, 32, 675–686. [Google Scholar] [CrossRef]
- Mookkan, M.; Nelson-Vasilchik, K.; Hague, J.; Kausch, A.; Zhang, Z.J. morphogenic regulator-mediated transformation of maize inbred B. Curr. Protoc. Plant Biol. 2018, 3, e20075. [Google Scholar] [CrossRef]
- Mookkan, M.; Nelson-Vasilchik, K.; Hague, J.; Zhang, Z.J.; Kausch, A.P. Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators baby boom and WUSCHEL. Plant Cell Rep. 2017, 36, 1477–1491. [Google Scholar] [CrossRef] [Green Version]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic regulators Baby boom and WUSCHEL improve monocot transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [Green Version]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, A.T. Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants 2019, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- El-Itriby, H.A.; Assem, S.K.; Hussein, E.H.A.; Abdel-Calil, F.M.; Madkour, M.A. Regeneration and transformation of Egyptian maize inbred lines via immature embryo culture and a biolistic particle delivery system. Vitr. Cell. Dev. Biol. Anim. 2003, 39, 524–531. [Google Scholar] [CrossRef]
- Frame, B.R.; Shou, H.; Chikwamba, R.K.; Zhang, Z.; Xiang, C.; Fonger, T.M.; Pegg, S.E.K.; Li, B.; Nettleton, D.S.; Pei, D.; et al. Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 2002, 129, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Frame, B.; Main, M.; Schick, R.; Wang, K. Genetic transformation using maize immature zygotic embryos. Methods Mol. Biol. 2010, 710, 327–341. [Google Scholar] [CrossRef]
- Gao, H.; Smith, J.; Yang, M.; Jones, S.; Djukanovic, V.; Nicholson, M.G.; West, A.; Bidney, D.; Falco, S.C.; Jantz, D.; et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2009, 61, 176–187. [Google Scholar] [CrossRef]
- Guan, Z.; Meng, X.; Sun, Z.; Xu, Z.; Song, R. Characterization of duplicated Dunaliella viridis SPT1 genes provides insights into early gene divergence after duplication. Gene 2008, 423, 36–42. [Google Scholar] [CrossRef]
- Wang, G.; Wang, G.; Zhang, X.; Wang, F.; Song, R. Isolation of high quality RNA from cereal seeds containing high levels of starch. Phytochem. Anal. 2012, 23, 159–163. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Anand, A.; Arling, M.L.; da Silva Conceicao, A.; Gordon-Kamm, W.J.; Hastings, C.E.; Hoerster, G.M.; Klein, T.M.; La Rota, C.M.; Lowe, K.S.; Tiwari, S.B. et al. Methods and Compositions for Rapid Plant Transformation. U.S. Patent US20170121722A1, 4 May 2017. [Google Scholar]
- Garnaat, C.; Lowe, K.S.; Roth, B. Zmaxig1-Specific Polynucleotides and Methods of Use. U.S. Patent WO2002006499A3, 13 July 2001. [Google Scholar]
- An, G. Development of plant promoter expression vectors and their use for analysis of differential activity of nopaline synthase promoter in transformed tobacco cells. Plant Physiol. 1986, 81, 86–91. [Google Scholar] [CrossRef] [Green Version]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.; Prasher, D. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [Green Version]
- Walters, D.A.; Vetsch, C.S.; Potts, D.E.; Lundquist, R.C. Transformation and inheritance of a hygromycin phosphotransferase gene in maize plants. Plant Mol. Biol. 1992, 18, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of maize. Nat. Protoc. 2007, 2, 1614–1621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenhard, M.; Jürgens, G.; Laux, T. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Ara-bidopsis shoot meristem regulation. Development 2002, 129, 3195–3206. [Google Scholar] [CrossRef]
- Bohorova, N.; Luna, B.; Brito, R.M.; Huerta, L.D. Regeneration potential of tropical, subtropical, midaltitude, and highland maize inbreds. Maydica 1995, 40, 275–281. [Google Scholar]
- Tomes, D.T.; Smith, O.S. The effect of parental genotype on initiation of embryogenic callus from elite maize (Zea mays L.) germplasm. Theor. Appl. Genet. 1985, 70, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Willman, M.R.; Schroll, S.M.; Hodges, T.K. Inheritance of somatic embryogenesis and plantlet regeneration from primary (type 1) callus in maize. Vitr. Cell. Dev. Biol. Anim. 1989, 25, 95–100. [Google Scholar] [CrossRef]
- Raihan, M.S.; Liu, J.; Huang, J.; Guo, H.; Pan, Q.; Yan, J. Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58 × SK maize population. Theor. Appl. Genet. 2016, 129, 1465–1477. [Google Scholar] [CrossRef]
- Raji, J.A.; Frame, B.; Little, D.; Santoso, T.J.; Wang, K.; Lagrimini, L.M. Agrobacterium- and biolistic-mediated transformation of maize b104 inbred. Adv. Struct. Saf. Stud. 2018, 1676, 15–40. [Google Scholar] [CrossRef]
- Deng, W.; Luo, K.; Li, Z.; Yang, Y. A novel method for induction of plant regeneration via somatic embryogenesis. Plant Sci. 2009, 177, 43–48. [Google Scholar] [CrossRef]
- Meyer, P.; Saedler, H. Homology-dependent gene silencing in plants. Annu. Rev. Plant Biol. 1996, 47, 23–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhi, L.; TeRonde, S.; Meyer, S.; Arling, M.L.; Iii, J.C.R.; Zhao, Z.-Y.; Jones, T.J.; Anand, A. Effect of Agrobacterium strain and plasmid copy number on transformation frequency, event quality and usable event quality in an elite maize cultivar. Plant Cell Rep. 2015, 34, 745–754. [Google Scholar] [CrossRef] [PubMed]






| Maize Inbred Lines | Vector | Number of Immature Embryos | Number of Positive T0 Plants | Transformation Ratio (%) |
|---|---|---|---|---|
| Zheng58 | pHB-CB-NW | 238 | 7 | 2.94 |
| pCA-GFP | ||||
| Zheng58 | pHB-CB-AW | 695 | 1 | 0.14 |
| pCA-GFP | ||||
| Mo17 | pHB-CB-AW | 301 | 2 | 0.66 |
| pCA-GFP | ||||
| Mo17 | pHB-PB-AW-GFP | 509 | 8 | 1.57 |
| Chang7-2 | data pHB-CB-AW-GFP | 692 | 4 | 0.58 |
| Maize Inbred Line | Event | Number of Detected T2 Seeds | Number of Coexistent Seeds | Number of Separated Seeds | Segregation Ratio (%) |
|---|---|---|---|---|---|
| Zheng58 | 1# | 59 | 55 | 4 | 6.78% |
| 2# | 107 | 78 | 4 | 2.80% | |
| 3# | 68 | 38 | 3 | 4.41% | |
| 4# | 148 | 118 | 4 | 2.03% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Y.; Chen, X.; Song, R.; Qi, W. Establishment of a Bivector Genetic Transformation System in Recalcitrant Maize Inbred Lines. Agriculture 2021, 11, 663. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture11070663
Gu Y, Chen X, Song R, Qi W. Establishment of a Bivector Genetic Transformation System in Recalcitrant Maize Inbred Lines. Agriculture. 2021; 11(7):663. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture11070663
Chicago/Turabian StyleGu, Yajing, Xuan Chen, Rentao Song, and Weiwei Qi. 2021. "Establishment of a Bivector Genetic Transformation System in Recalcitrant Maize Inbred Lines" Agriculture 11, no. 7: 663. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture11070663