Impacts of Background Removal on Convolutional Neural Networks for Plant Disease Classification In-Situ
Abstract
:1. Introduction
1.1. Related Works
1.2. Problems with a Cluttered Background
2. Materials and Methods
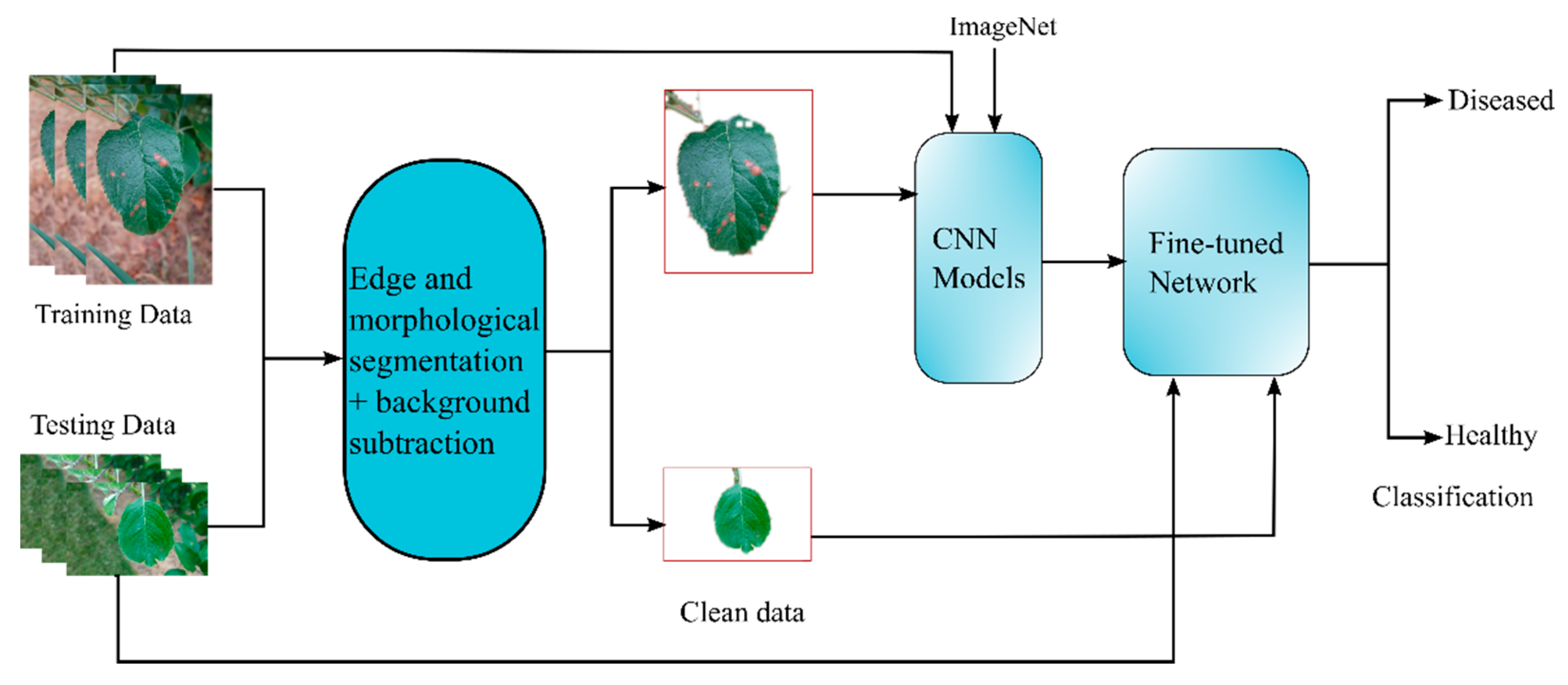
2.1. Proposed Approach
2.2. Edge and Morphological Based Segmentation
| Algorithm 1. Canny edge detection algorithm |
|
| Algorithm 2. Algorithm for Background Removal |
|
2.3. Background Subtraction
2.4. Transfer Learning and Fine-Tuning
2.5. Convolutional Block
2.6. Dataset
3. Results and Discussion
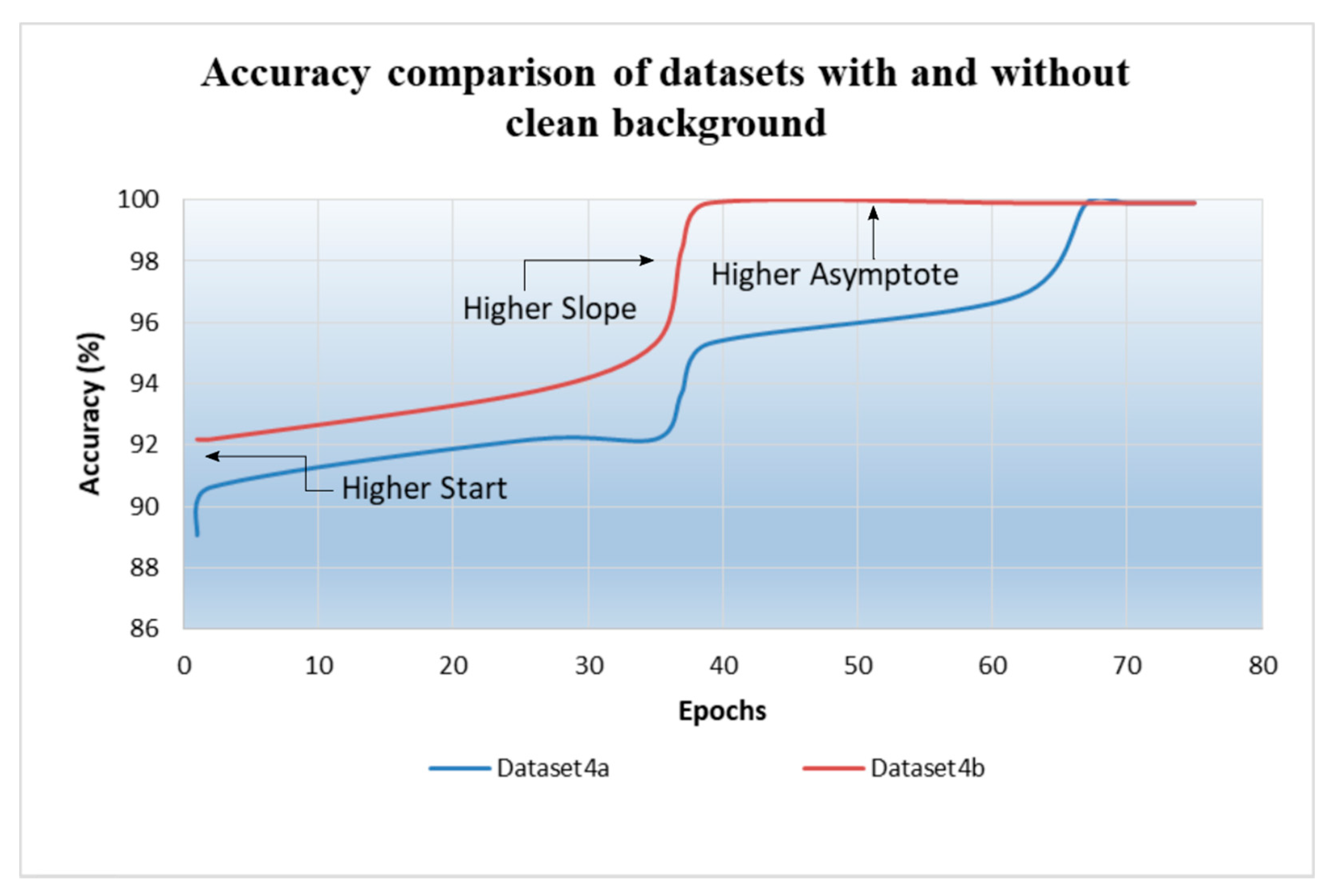
3.1. Background Removal
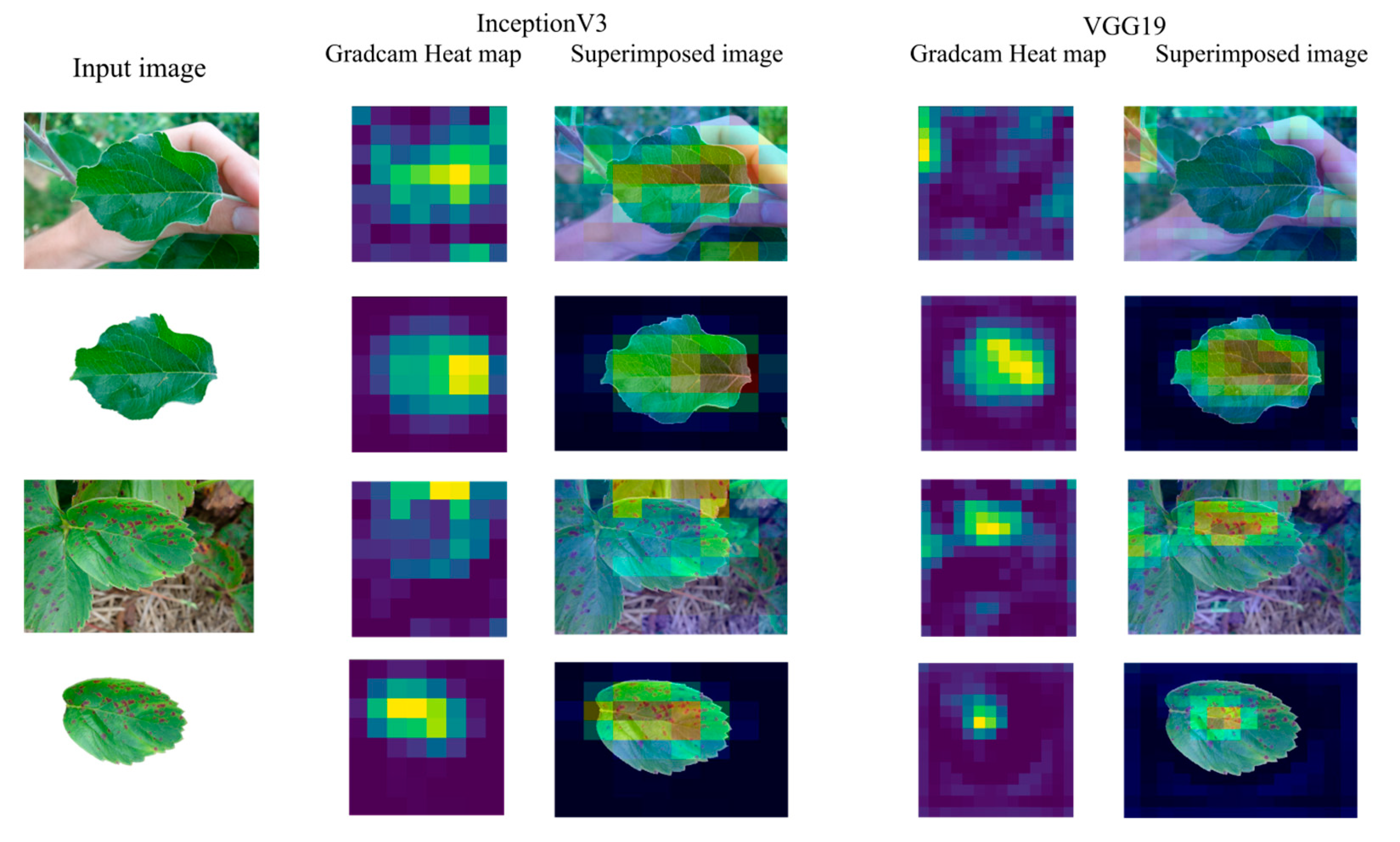
3.2. Grad-CAM Class Activation Visualization
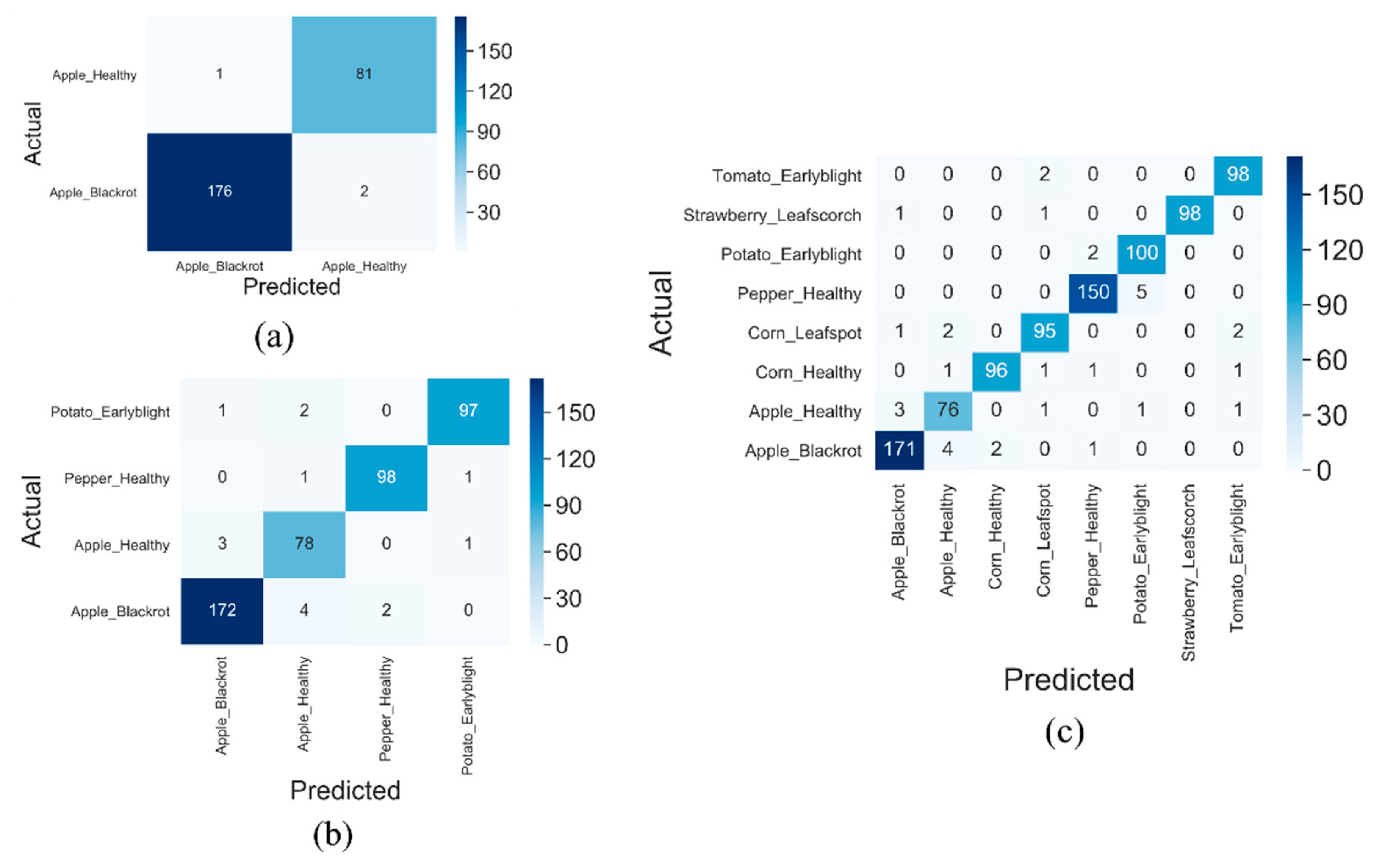
3.3. Classification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Gonzalez, A.N.; Villamizar, M.; Canevet, O.; Odobez, J.-M. Efficient Convolutional Neural Networks for Depth-Based Multi-Person Pose Estimation. IEEE Trans. Circ. Syst. Video Technol. 2020, 30, 4207–4221. [Google Scholar] [CrossRef] [Green Version]
- Simard, P.; Steinkraus, D.; Platt, J. Best practices for convolutional neural networks applied to visual document analysis. In Proceedings of the 7th International Conference on Document Analysis and Recognition, Edinburgh, UK, 3–6 August 2003; pp. 958–963. [Google Scholar]
- Li, Q.; Cai, W.; Wang, X.; Zhou, Y.; Feng, D.D.; Chen, M. Medical image classification with convolutional neural network. In Proceedings of the 2014 13th International Conference on Control Automation Robotics & Vision (ICARCV), Singapore, 10–12 December 2014; pp. 844–848. [Google Scholar]
- Kc, K.; Yin, Z.; Wu, M.; Wu, Z. Evaluation of deep learning-based approaches for COVID-19 classification based on chest X-ray images. Signal Image Video Process. 2021, 15, 959–966. [Google Scholar] [CrossRef]
- Zhao, K.; Kang, J.; Jung, J.; Sohn, G. Building extraction from satellite images using mask R-CNN with building bound-ary regularization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; pp. 247–251. [Google Scholar]
- Amit, S.N.K.B.; Shiraishi, S.; Inoshita, T.; Aoki, Y. Analysis of satellite images for disaster detection. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 5189–5192. [Google Scholar]
- Yuan, Y.; Zheng, X.; Lu, X. Hyperspectral Image Superresolution by Transfer Learning. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 1963–1974. [Google Scholar] [CrossRef]
- Mei, S.; Ji, J.; Hou, J.; Li, X.; Du, Q. Learning Sensor-Specific Spatial-Spectral Features of Hyperspectral Images via Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4520–4533. [Google Scholar] [CrossRef]
- Rajnoha, M.; Burget, R.; Povoda, L. Image Background Noise Impact on Convolutional Neural Network Training. In Proceedings of the 2018 10th International Congress on Ultra Modern Telecommunications and Control Systems and Workshops (ICUMT), Moscow, Russia, 5–9 November 2018; pp. 1–4. [Google Scholar]
- Singh, V.; Varsha Misra, A.K. Detection of unhealthy region of plant leaves using image processing and genetic algo-rithm. In Proceedings of the 2015 International Conference on Advances in Computer Engineering and Applications, Ghaziabad, India, 19–20 March 2015; pp. 1028–1032. [Google Scholar]
- Setitra, I.; Larabi, S. Background Subtraction Algorithms with Post-processing: A Review. In Proceedings of the 2014 22nd International Conference on Pattern Recognition, Stockholm, Sweden, 24–28 August 2014; pp. 2436–2441. [Google Scholar]
- Wang, K.; Gao, X.; Zhao, Y.; Li, X.; Dou, D.; Xu, C.-Z. Pay attention to features, transfer learn faster CNNs. In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How Transferable Are Features in Deep Neural Networks? arXiv 2014, arXiv:1411.1792. [Google Scholar]
- Salman, H.; Ilyas, A.; Engstrom, L.; Kapoor, A.; Madry, A. Do adversarially robust imagenet models transfer better? arXiv 2020, arXiv:2007.08489. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arivazhagan, S.; Shebiah, R.N.; Ananthi, S.; Varthini, S.V. Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric. Eng. Int. CIGR J. 2013, 15, 211–217. [Google Scholar]
- Singh, V.; Misra, A. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Al Bashish, D.; Braik, M.; Bani-Ahmad, S. Detection and classification of leaf diseases using K-means-based segmentation and. Inf. Technol. J. 2011, 10, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, K.; Yang, F.; Pan, S.; Han, Y. Image segmentation of overlapping leaves based on Chan–Vese model and Sobel operator. Inf. Process. Agric. 2018, 5, 1–10. [Google Scholar] [CrossRef]
- Chen, C.-T.; Su, C.-Y.; Kao, W.-C. An enhanced segmentation on vision-based shadow removal for vehicle detection. In Proceedings of the the 2010 International Conference on Green Circuits and Systems, Shanghai, China, 21–23 June 2010; pp. 679–682. [Google Scholar]
- Jamal, I.; Akram, M.U.; Tariq, A. Retinal Image Preprocessing: Background and Noise Segmentation. TELKOMNIKA (Telecommun. Comput. Electron. Control. 2012, 10, 537–544. [Google Scholar] [CrossRef]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Kc, K.; Yin, Z.; Wu, M.; Wu, Z. Depthwise separable convolution architectures for plant disease classification. Comput. Electron. Agric. 2019, 165, 104948. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.F. Imagenet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Kamal, K.; Yin, Z.; Li, B.; Ma, B.; Wu, M. Transfer Learning for Fine-Grained Crop Disease Classification Based on Leaf Images. In Proceedings of the 2019 10th Workshop on Hyperspectral Imaging and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 24–26 September 2019. [Google Scholar]
- Chen, J.; Chen, J.; Zhang, D.; Sun, Y.; Nanehkaran, Y. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. 2020, 173, 105393. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Comput. Electron. Agric. 2018, 153, 46–53. [Google Scholar] [CrossRef]
- Nayak, A.A.; Venugopala, P.S.; Sarojadevi, H.; Chiplunkar, N.N. An approach to improvise canny edge detection using morphological filters. Int. J. Comput. Appl. 2015, 116, 38–42. [Google Scholar]
- Mandellos, N.A.; Keramitsoglou, I.; Kiranoudis, C.T. A background subtraction algorithm for detecting and tracking vehicles. Expert Syst. Appl. 2011, 38, 1619–1631. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Kaiming, H.; Xiangyu, Z.; Shaoqing, R.; Jian, S. Deep Learning with Depthwise Separable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef] [Green Version]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Con-ference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; p. 18311827. [Google Scholar]
- Hughes, D.P.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics through machine learning and crowdsourcing. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef] [Green Version]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Wang, G.; Sun, Y.; Wang, J. Automatic Image-Based Plant Disease Severity Estimation Using Deep Learning. Comput. Intell. Neurosci. 2017, 2017, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Hyperparameters | Data Augmentation | |
|---|---|---|
| Optimizer | Nadam | Rescaling |
| Learning rate | Horizontal flip | |
| Exponential decay rate () | 0.9 | Zoom range = 0.3 |
| Exponential decay rate () | 0.999 | Width shift range = 0.3 |
| Epsilon (ε) | Height shift range = 0.3 | |
| Batch size | 128 | Rotation range = 30 |
| Maximum epoch | 500 | Zca whitening |
| Activation function | Rectified Linear Unit (ReLU) | Zca epsilon = |
| Dataset1b | Dataset2b | Dataset3b | ||
|---|---|---|---|---|
| Metrics | DenseNet121 | InceptionV3 | DenseNet121 | |
| Models | ||||
| Precision | 0.9944 | 0.9829 | 0.9639 | |
| Recall | 0.9888 | 0.9773 | 0.9524 | |
| Specificity | 0.9878 | 0.963 | 0.9240 | |
| F1-score | 0.9916 | 0.98 | 0.958 | |
| Accuracy (%) | 98.9 | 96.7 | 93.57 | |
| Related Work | Accuracy (%) | Classes | Total Images | Train-Test Split (%) | Model |
|---|---|---|---|---|---|
| Mohanty et al. [17] | 31.4 | - | 121 | 80–20 | GoogLeNet |
| Ferentinos et al. [41] | 33.27 | 12 | - | 55.8–44.2 | VGG |
| Kamal et al. [25] | 36.03 | 11 | 22110 | 48.1–51.9 | MobileNet |
| Wang et al. [42] | 90.4 | 4 | 2086 | 80–20 | Fine-tuned InceptionV3 |
| Proposed system (Cluttered background) | 87.12 | 2 | 1262 | 80–20 | Fine-tuned Densenet121 |
| Proposed system (Background removed) | 93.57 | 8 | 4558 | 80–20 | Fine-tuned Densenet121 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
KC, K.; Yin, Z.; Li, D.; Wu, Z. Impacts of Background Removal on Convolutional Neural Networks for Plant Disease Classification In-Situ. Agriculture 2021, 11, 827. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture11090827
KC K, Yin Z, Li D, Wu Z. Impacts of Background Removal on Convolutional Neural Networks for Plant Disease Classification In-Situ. Agriculture. 2021; 11(9):827. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture11090827
Chicago/Turabian StyleKC, Kamal, Zhendong Yin, Dasen Li, and Zhilu Wu. 2021. "Impacts of Background Removal on Convolutional Neural Networks for Plant Disease Classification In-Situ" Agriculture 11, no. 9: 827. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture11090827






