A Label-Free Electrochemical Impedance Genosensor Coupled with Recombinase Polymerase Amplification for Genetically Modified Maize Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Apparatus
2.2. Preparation of GCDs
2.3. DNA Extraction
2.4. Primer Design and RPA Reactions
2.5. Label-Free EI Genosensor for GM Crop Detection
3. Results and Discussion
3.1. Characterization of GCDs-Based EI Genosensor
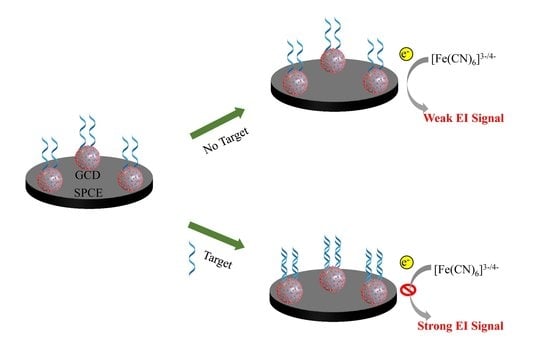
3.2. Principle of Label-Free EI Genosensor
3.3. Optimization of Experimental Conditions
3.4. Specificity, Repeatability and Stability of EI Genosensor
3.5. Sensitivity of EI Genosensor
3.6. Identification of GM Maize in Entire Process
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- ISAAA. Global Status of Commercialized Biotech/GM Crops in 2019: Biotech Crops Drive Socio-Economic Development and Sustainable Environment in the New Frontier; ISAAA Briefs NO. 55; ISAAA: Ithaca, NY, USA, 2019. [Google Scholar]
- Cottenet, G.; Blancpain, C.; Sonnard, V.; Chuah, P.F. Development and validation of a multiplex real-time PCR method to simultaneously detect 47 targets for the identification of genetically modified organisms. Anal. Bioanal. Chem. 2013, 405, 6831–6844. [Google Scholar] [CrossRef] [PubMed]
- Li, X.F.; Wu, Y.H.; Li, J.; Li, Y.J.; Long, L.K.; Li, F.W.; Wu, G. Development and Validation of A 48-Target Analytical Method for High-throughput Monitoring of Genetically Modified Organisms. Sci. Rep. 2015, 5, 7616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosa, S.F.; Gatto, F.; Angers-Loustau, A.; Petrillo, M.; Kreysa, J.; Querci, M. Development and applicability of a ready-to-use PCR system for GMO screening. Food Chem. 2016, 201, 110–119. [Google Scholar] [CrossRef]
- Rao, J.; Yang, L.; Guo, J.; Quan, S.; Chen, G.; Zhao, X.; Zhang, D.; Shi, J. Development of event-specific qualitative and quantitative PCR detection methods for the transgenic maize BVLA430101. Eur. Food Res. Technol. 2016, 242, 1277–1284. [Google Scholar] [CrossRef]
- Yang, L.T.; Yang, Y.; Jin, W.J.; Zhang, X.J.; Li, X.Y.; Wu, Y.H.; Li, J.; Li, L. Development and Interlaboratories Validation of Event-Specific Quantitative Real-Time PCR Method for Genetically Modified Rice G6H1 Event. J. Agric. Food Chem. 2018, 66, 8179–8186. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J. Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef]
- Lutz, S.; Weber, P.; Focke, M.; Faltin, B.; Hoffmann, J.; Müller, C.; Mark, D.; Roth, G.; Munday, P.; Armes, N.; et al. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA). Lab. Chip. 2010, 10, 887–893. [Google Scholar] [CrossRef]
- Xu, Y.; Wei, Y.; Cheng, N.; Huang, K.; Wang, W.; Zhang, L.; Xu, W.; Luo, Y. Nucleic Acid Biosensor Synthesis of an All-in-One Universal Blocking Linker Recombinase Polymerase Amplification with a Peptide Nucleic Acid-Based Lateral Flow Device for Ultrasensitive Detection of Food Pathogens. Anal. Chem. 2018, 90, 708–715. [Google Scholar] [CrossRef]
- Hu, M.; Yuan, C.; Tian, T.; Wang, X.; Sun, J.; Xiong, E.; Zhou, X. Single-Step, Salt-Aging-Free, and Thiol-Free Freezing Construction of AuNP-Based Bioprobes for Advancing CRISPR-Based Diagnostics. J. Am. Chem. Soc. 2020, 142, 7506–7513. [Google Scholar] [CrossRef]
- Wang, X.; Xiong, E.; Tian, T.; Cheng, M.; Lin, W.; Wang, H.; Zhang, G.; Sun, J.; Zhou, X. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9-Mediated Lateral Flow Nucleic Acid Assay. ACS Nano 2020, 14, 2497–2508. [Google Scholar] [CrossRef]
- An, N.; Li, K.; Zhang, Y.; Wen, T.; Liu, W.; Liu, G.; Li, L.; Jin, W. A multiplex and regenerable surface plasmon resonance (MR-SPR) biosensor for DNA detection of genetically modified organisms. Talanta 2021, 231, 122361. [Google Scholar] [CrossRef] [PubMed]
- Mannelli, I.; Minunni, M.; Tombelli, S.; Mascini, M. Quartz crystal microbalance (QCM) affinity biosensor for genetically modified organisms (GMOs) detection. Biosens. Bioelectron. 2003, 18, 129–140. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, M.; Ma, M.; Hai, H.; Li, J.; Shan, Y. A novel electrochemical DNA biosensor for transgenic soybean detection based on triple signal amplification. Anal. Chim. Acta 2019, 1078, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zheng, L.; Chen, Y.; Xue, F.; Cheng, L.; Adeloju, S.B.; Chen, W. A novel GMO biosensor for rapid ultrasensitive and simultaneous detection of multiple DNA components in GMO products. Biosens. Bioelectron. 2015, 66, 431–437. [Google Scholar] [CrossRef]
- Placido, A.; Pereira, C.; Guedes, A.; Fatima Barroso, M.; Miranda-Castro, R.; De-los-Santos-Alvarezd, N.; Delerue-Matos, C. Electrochemical genoassays on gold-coated magnetic nanoparticles to quantify genetically modified organisms (GMOs) in food and feed as GMO percentage. Biosens. Bioelectron. 2018, 110, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Sun, L.; Liu, Q.; Han, E.; Hao, N.; Zhang, L.; Wang, S.; Cai, J.; Wang, K. Photoelectrochemical CaMV35S biosensor for discriminating transgenic from non-transgenic soybean based on SiO2@CdTe quantum dots core-shell nanoparticles as signal indicators. Talanta 2016, 161, 211–218. [Google Scholar] [CrossRef]
- Zhu, D.; Liu, J.; Tang, Y.; Xing, D. A reusable DNA biosensor for the detection of genetically modified organism using magnetic bead-based electrochemiluminescence. Sens. Actuators B Chem. 2010, 149, 221–225. [Google Scholar] [CrossRef]
- Bonanni, A.; Esplandiu, M.J.; Del Valle, M. Impedimetric genosensors employing COOH-modified carbon nanotube screen-printed electrodes. Biosens. Bioelectron. 2009, 24, 2885–2891. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Q.; Li, H.; Li, Y.; Hao, N.; Qian, J.; Zhu, W.; Wang, K. Fabrication of label-free electrochemical impedimetric DNA biosensor for detection of genetically modified soybean by recognizing CaMV 35S promoter. J. Electroanal. Chem. 2016, 782, 19–25. [Google Scholar] [CrossRef]
- Bolat, G.; Yaman, Y.T.; Abaci, S. Molecularly imprinted electrochemical impedance sensor for sensitive dibutyl phthalate (DBP) determination. Sens. Actuators B Chem. 2019, 299, 127000. [Google Scholar] [CrossRef]
- Mousavisani, S.Z.; Raoof, J.B.; Turner, A.; Ojani, R.; Mak, W.C. Label-free DNA sensor based on diazonium immobilisation for detection of DNA damage in breast cancer 1 gene. Sens. Actuators B Chem. 2018, 264, 59–66. [Google Scholar] [CrossRef]
- Aghili, Z.; Nasirizadeh, N.; Divsalar, A.; Shoeibi, S.; Yaghmaei, P. A nanobiosensor composed of Exfoliated Graphene Oxide and Gold Nano-Urchins for detection of GMO products. Biosens. Bioelectron. 2017, 95, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Fortunati, S.; Rozzi, A.; Curti, F.; Giannetto, M.; Corradini, R.; Careri, M. Novel amperometric genosensor based on peptide nucleic acid (PNA) probes immobilized on carbon nanotubes-screen printed electrodes for the determination of trace levels of non-amplified DNA in genetically modified (GM) soy. Biosens. Bioelectron. 2019, 129, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Zhou, Y.; Leblanc, R.M.; Peng, Z. Recent Developments of Carbon Dots in Biosensing: A Review. ACS Sens. 2020, 5, 2724–2741. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Hua, X.; Wang, Z.; Yang, X.; Huang, H. Real-Time Imaging Redox Status in Biothiols and Ferric Metabolism of Cancer Cells in Ferroptosis Based on Switched Fluorescence Response of Gold Carbon Dots. Anal. Chem. 2020, 92, 11420–11428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, D.; Huang, H.; Liu, L.; Zhou, Y.; Xia, X.; Deng, K.; Liu, X. Preparation of Gold-Carbon Dots and Ratiometric Fluorescence Cellular Imaging. ACS Appl. Mater. Interfaces 2016, 8, 6646–6655. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, L.; Zhang, L.; Zhang, X.; Li, X.; Zhai, S.; Gao, H.; Li, Y.; Wu, G.; Wu, Y. Development of a certified genomic DNA reference material for detection and quantification of genetically modified rice KMD. Anal. Bioanal. Chem. 2020, 412, 7007–7016. [Google Scholar] [CrossRef]
- Arumuganathan, K.; Earle, E.D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Hou, J.; Dong, J.; Zhu, H.; Teng, X.; Ai, S.; Mang, M. A simple and sensitive fluorescent sensor for methyl parathion based on L-tyrosine methyl ester functionalized carbon dots. Biosens. Bioelectron. 2015, 68, 20–26. [Google Scholar] [CrossRef]
- Kalogianni, D.P.; Koraki, T.; Christopoulos, T.K.; Ioannou, P.C. Nanoparticle-based DNA biosensor for visual detection of genetically modified organisms. Biosens. Bioelectron. 2006, 21, 1069–1076. [Google Scholar] [CrossRef]
- Cheng, N.; Shang, Y.; Xu, Y.; Zhang, L.; Luo, Y.; Huang, K.; Xu, W. On-site detection of stacked genetically modified soybean based on event specific TM-LAMP and a DNAzyme-lateral flow biosensor. Biosens. Bioelectron. 2017, 91, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Luo, Y.; Huang, K.; Yang, Z.; Wan, Y.; Xu, W. Single universal primer recombinase polymerase amplification-based lateral flow biosensor (SUP-RPA-LFB) for multiplex detection of genetically modified maize. Anal. Chim. Acta 2020, 1127, 217–224. [Google Scholar] [CrossRef] [PubMed]





| Method | Analyte | LOD | Reference |
|---|---|---|---|
| Qualitative PCR | Rapeseed MS1 | 2.5 copies/μL | [5] |
| Rapeseed RF1 | 50 copies/μL | ||
| Rapeseed RF2 | 5.0 copies/μL | ||
| Real-time PCR | Rice G6H1 | 10 copies/μL | [6] |
| Multiplex real-time PCR | 9 taxon-, 10 element- and 28 event-specific targets | 1.0–16 copies/μL | [2] |
| Ready-to-use PCR plates | 7 taxon-, 5 element-, 1 construct- and 3 event-specific targets | 5.0–20 copies/μL | [4] |
| Electrochemical biosensor | GM soybean and rice | 0.10% GM content | [15] |
| Forster resonance energy transfer assay | CaMV35s | 0.50% GM content | [17] |
| Dry-reagent DNA biosensor | CaMV35s and NOS | 0.10% GM content | [31] |
| DNAzyme-lateral flow biosensor | DP305423 × GTS 40-3-2 soybean | 0.10% GM content | [32] |
| SUP-RPA-lateral flow biosensor | MON810, MON863, MON89034 | 50 copies/μL | [33] |
| RPA-based EI genosensor | Maize Ruifeng 12-5 | 0.10% GM content (36 copies/μL) | The proposed method |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, D.; Zhai, S.; Yang, Y.; Wu, Y.; Li, J.; Yan, X.; Shen, P.; Gao, H.; Wu, G. A Label-Free Electrochemical Impedance Genosensor Coupled with Recombinase Polymerase Amplification for Genetically Modified Maize Detection. Agriculture 2022, 12, 454. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture12040454
Cui D, Zhai S, Yang Y, Wu Y, Li J, Yan X, Shen P, Gao H, Wu G. A Label-Free Electrochemical Impedance Genosensor Coupled with Recombinase Polymerase Amplification for Genetically Modified Maize Detection. Agriculture. 2022; 12(4):454. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture12040454
Chicago/Turabian StyleCui, Dandan, Shanshan Zhai, Yao Yang, Yuhua Wu, Jun Li, Xiaohong Yan, Ping Shen, Hongfei Gao, and Gang Wu. 2022. "A Label-Free Electrochemical Impedance Genosensor Coupled with Recombinase Polymerase Amplification for Genetically Modified Maize Detection" Agriculture 12, no. 4: 454. https://0-doi-org.brum.beds.ac.uk/10.3390/agriculture12040454