Aptamer Conjugated Indium Phosphide Quantum Dots with a Zinc Sulphide Shell as Photoluminescent Labels for Acinetobacter baumannii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of InP/ZnS Quantum Dots
2.2. Ligand Exchange with Mercaptosuccinic Acid
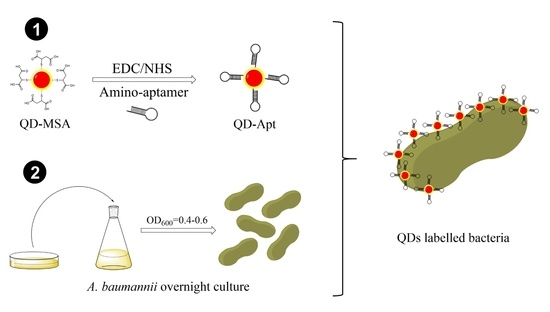
2.3. Conjugation of the InP/ZnS QDs-MSA with Aptamers
2.4. Bacterial Culture and QDs-Aptamer Coupling
2.5. Calibration Curve
2.6. Transmission Electron Microscopy
2.7. Photoluminescence Spectroscopy
2.8. Confocal Microscopy
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, X.; Hilliard, L.R.; Mechery, S.J.; Wang, Y.; Bagwe, R.P.; Jin, S.; Tan, W. A rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticles. Proc. Natl. Acad. Sci. USA 2004, 101, 15027–15032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter Baumannii and Pseudomonas Aeruginosa in Health Care Facilities. Available online: https://www.who.int/publications/i/item/guidelines-for-the-prevention-and-control-of-carbapenem-resistant-enterobacteriaceae-acinetobacter-baumannii-and-pseudomonas-aeruginosa-in-health-care-facilities (accessed on 10 June 2021).
- Fournier, P.E.; Richet, H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 2006, 42, 692–699. [Google Scholar] [CrossRef] [Green Version]
- Anstey, N.M.; Currie, B.J.; Withnall, K.M. Community-acquired acinetobacter pneumonia in the Northern Territory of Australia. Clin. Infect. Dis. 1992, 14, 83–91. [Google Scholar] [CrossRef]
- Chu, Y.W.; Leung, C.M.; Houang, E.T.S.; Ng, K.C.; Leung, C.B.; Leung, H.Y.; Cheng, A.F.B. Skin carriage of acinetobacters in Hong Kong. J. Clin. Microbiol. 1999, 37, 2962–2967. [Google Scholar] [CrossRef] [Green Version]
- Wenzler, E.; Goff, D.A.; Mangino, J.E.; Reed, E.E.; Wehr, A.; Bauer, K.A. Impact of rapid identification of Acinetobacter Baumannii via matrix-assisted laser desorption ionization time-of-flight mass spectrometry combined with antimicrobial stewardship in patients with pneumonia and/or bacteremia. Diagn. Microbiol. Infect. Dis. 2016, 84, 63–68. [Google Scholar] [CrossRef]
- Kempf, M.; Rolain, J.M. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: Clinical impact and therapeutic options. Int. J. Antimicrob. Agents 2012, 39, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Kempf, M.; Abdissa, A.; Diatta, G.; Trape, J.F.; Angelakis, E.; Mediannikov, O.; La Scola, B.; Raoult, D. Detection of Acinetobacter baumannii in human head and body lice from Ethiopia and identification of new genotypes. Int. J. Infect. Dis. 2012, 16, e680–e683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Towner, K.J. Acinetobacter: An old friend, but a new enemy. J. Hosp. Infect. 2009, 73, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Garnacho-Montero, J.; Dimopoulos, G.; Poulakou, G.; Akova, M.; Cisneros, J.M.; De Waele, J.; Petrosillo, N.; Seifert, H.; Timsit, J.F.; Vila, J.; et al. Task force on management and prevention of Acinetobacter baumannii infections in the ICU. Intensive Care Med. 2015, 41, 2057–2075. [Google Scholar] [CrossRef] [PubMed]
- Soo, P.C.; Tseng, C.C.; Ling, S.R.; Liou, M.L.; Liu, C.C.; Chao, H.J.; Lin, T.Y.; Chang, K.C. Rapid and sensitive detection of Acinetobacter baumannii using loop-mediated isothermal amplification. J. Microbiol. Methods 2013, 92, 197–200. [Google Scholar] [CrossRef]
- Turton, J.F.; Woodford, N.; Glover, J.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Identification of Acinetobacter baumannii by detection of the bla OXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006, 44, 2974–2976. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.Y.; Hopping, G.C.; Vaidyanathan, U.; Ronquillo, Y.C.; Hoopes, P.C.; Moshirfar, M. Polymerase Chain Reaction and Its Application in the Diagnosis of Infectious Keratitis. Med. Hypothesis Discov. Innov. Ophthalmol. J. 2019, 8, 152–155. [Google Scholar]
- Acquah, C.; Danquah, M.K.; Yon, J.L.S.; Sidhu, A.; Ongkudon, C.M. A review on immobilised aptamers for high throughput biomolecular detection and screening. Anal. Chim. Acta 2015, 888, 10–18. [Google Scholar] [CrossRef] [Green Version]
- Rasoulinejad, S.; Gargari, S.L.M. Aptamer-nanobody based ELASA for specific detection of Acinetobacter baumannii isolates. J. Biotechnol. 2016, 231, 46–54. [Google Scholar] [CrossRef]
- Dunn, M.R.; Jimenez, R.M.; Chaput, J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017, 1, 0076. [Google Scholar] [CrossRef]
- Ikanovic, M.; Rudzinski, W.E.; Bruno, J.G.; Allman, A.; Carrillo, M.P.; Dwarakanath, S.; Bhahdigadi, S.; Rao, P.; Kiel, J.L.; Andrews, C.J. Fluorescence assay based on aptamer-quantum dot binding to bacillus thuringiensis spores. J. Fluoresc. 2007, 17, 193–199. [Google Scholar] [CrossRef]
- Bilan, R.; Nabiev, I.; Sukhanova, A. Quantum Dot-Based Nanotools for Bioimaging, Diagnostics, and Drug Delivery. ChemBioChem 2016, 17, 2103–2114. [Google Scholar] [CrossRef]
- Su, C.H.; Tsai, M.H.; Lin, C.Y.; Ma, Y.D.; Wang, C.H.; Chung, Y.D.; Lee, G.B. Dual aptamer assay for detection of Acinetobacter baumannii on an electromagnetically-driven microfluidic platform. Biosens. Bioelectron. 2020, 159, 112148. [Google Scholar] [CrossRef]
- Wen, L.; Qiu, L.; Wu, Y.; Hu, X.; Zhang, X. Aptamer-modified semiconductor quantum dots for biosensing applications. Sensors 2017, 17, 1736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayupova, D.; Dobhal, G.; Laufersky, G.; Nann, T.; Goreham, R.V. An in vitro investigation of cytotoxic effects of InP/ZnS quantum dots with different surface chemistries. Nanomaterials 2019, 9, 135. [Google Scholar] [CrossRef] [Green Version]
- Brunetti, V.; Chibli, H.; Fiammengo, R.; Galeone, A.; Malvindi, M.A.; Vecchio, G.; Cingolani, R.; Nadeau, J.L.; Pompa, P.P. InP/ZnS as a safer alternative to CdSe/ZnS core/shell quantum dots: In vitro and in vivo toxicity assessment. Nanoscale 2013, 5, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.T.; Goldman, E.R.; Anderson, G.P.; Mauro, J.M.; Mattoussi, H. Use of Luminescent CdSe-ZnS Nanocrystal Bioconjugates in Quantum Dot-Based Nanosensors. Phys. Status Solidi (B) 2002, 229, 427–432. [Google Scholar] [CrossRef]
- Kloepfer, J.A.; Mielke, R.E.; Wong, M.S.; Nealson, K.H.; Stucky, G.; Nadeau, J.L. Quantum dots as strain- and metabolism-specific microbiological labels. Appl. Environ. Microbiol. 2003, 69, 4205–4213. [Google Scholar] [CrossRef] [Green Version]
- Tessier, M.D.; Dupont, D.; De Nolf, K.; De Roo, J.; Hens, Z. Economic and Size-Tunable Synthesis of InP/ZnE (E = S, Se) Colloidal Quantum Dots. Chem. Mater. 2015, 27, 4893–4898. [Google Scholar] [CrossRef] [Green Version]
- Yong, K.T.; Ding, H.; Roy, I.; Law, W.C.; Bergey, E.J.; Maitra, A.; Prasad, P.N. Imaging pancreatic cancer using bioconjugated inp quantum dots. ACS Nano 2009, 3, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.H.; Yu, H.-W.; Kim, Y.-H.; Kim, I.S.; Jang, A. Potential of Fluorophore Labeled Aptamers for Pseudomonas aeruginosa Detection in Drinking Water. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 171. [Google Scholar] [CrossRef]
- Kloepfer, J.A.; Mielke, R.E.; Nadeau, J.L. Uptake of CdSe and CdSe/ZnS quantum dots into bacteria via purine-dependent mechanisms. Appl. Environ. Microbiol. 2005, 71, 2548–2557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruno, J.G. Application of DNA aptamers and quantum dots to lateral flow test strips for detection of foodborne pathogens with improved sensitivity versus colloidal gold. Pathogens 2014, 3, 341–355. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, X.P. Fabrication of vascular endothelial growth factor antibody bioconjugated ultrasmall near-infrared fluorescent Ag2S quantum dots for targeted cancer imaging in vivo. Chem. Commun. 2013, 49, 3324–3326. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y. Simultaneous detection of Escherichia coli O157:H7 and Salmonella Typhimurium using quantum dots as fluorescence labels. Analyst 2006, 131, 394–401. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.F. Probing the interactions of chitosan capped CdS quantum dots with pathogenic bacteria and their biosensing application. J. Mater. Chem. B 2013, 1, 6094–6106. [Google Scholar] [CrossRef]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L.V. Aptamers: Problems, solutions and prospects. Acta Nat. 2013, 5, 34–43. [Google Scholar] [CrossRef] [Green Version]
- Song, K.M.; Jeong, E.; Jeon, W.; Cho, M.; Ban, C. Aptasensor for ampicillin using gold nanoparticle based dual fluorescence-colorimetric methods. Anal. Bioanal. Chem. 2012, 402, 2153–2161. [Google Scholar] [CrossRef] [PubMed]
- Shannon Pendergrast, P.; Nicholas Marsh, H.; Grate, D.; Healy, J.M.; Stanton, M. Nucleic acid aptamers for target validation and therapeutic applications. J. Biomol. Tech. 2005, 16, 224–234. [Google Scholar]
- Mann, D.; Reinemann, C.; Stoltenburg, R.; Strehlitz, B. In vitro selection of DNA aptamers binding ethanolamine. Biochem. Biophys. Res. Commun. 2005, 338, 1928–1934. [Google Scholar] [CrossRef] [PubMed]
- Dickey, D.D.; Giangrande, P.H. Oligonucleotide aptamers: A next-generation technology for the capture and detection of circulating tumor cells. Methods 2016, 97, 94–103. [Google Scholar] [CrossRef] [Green Version]
- Sark, W.V.; Frederix, P.L.; Bol, A.A.; Gerritsen, H.C.; Meijerink, A. Blueing, Bleaching, and Blinking of Single CdSe/ZnS Quantum Dots. J. Phys. Chem. A 2002, 3, 871–879. [Google Scholar]
- Stoeckenius, W. Electron microscopy of DNA molecules “stained” with heavy metal salts. J. Biophys. Biochem. Cytol. 1961, 11, 297–310. [Google Scholar] [CrossRef] [Green Version]
- Karami-Zarandi, M.; Douraghi, M.; Vaziri, B.; Adibhesami, H.; Rahbar, M.; Yaseri, M. Variable spontaneous mutation rate in clinical strains of multidrug-resistant Acinetobacter baumannii and differentially expressed proteins in a hypermutator strain. Mutat. Res.-Fundam. Mol. Mech. Mutagen. 2017, 800–802, 37–45. [Google Scholar] [CrossRef]
- Nakano, K.; Nomura, R.; Shimizu, N.; Nakagawa, I.; Hamada, S.; Ooshima, T. Development of a PCR method for rapid identification of new Streptococcus mutans serotype k strains. J. Clin. Microbiol. 2004, 42, 4925–4930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Helden, P.D. Bacterial genetics and strain variation. Novartis Found. Symp. 1998, 217, 178–194. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayed, Z.; Malhotra, S.; Dobhal, G.; Goreham, R.V. Aptamer Conjugated Indium Phosphide Quantum Dots with a Zinc Sulphide Shell as Photoluminescent Labels for Acinetobacter baumannii. Nanomaterials 2021, 11, 3317. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11123317
Ayed Z, Malhotra S, Dobhal G, Goreham RV. Aptamer Conjugated Indium Phosphide Quantum Dots with a Zinc Sulphide Shell as Photoluminescent Labels for Acinetobacter baumannii. Nanomaterials. 2021; 11(12):3317. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11123317
Chicago/Turabian StyleAyed, Zeineb, Shiana Malhotra, Garima Dobhal, and Renee V. Goreham. 2021. "Aptamer Conjugated Indium Phosphide Quantum Dots with a Zinc Sulphide Shell as Photoluminescent Labels for Acinetobacter baumannii" Nanomaterials 11, no. 12: 3317. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11123317