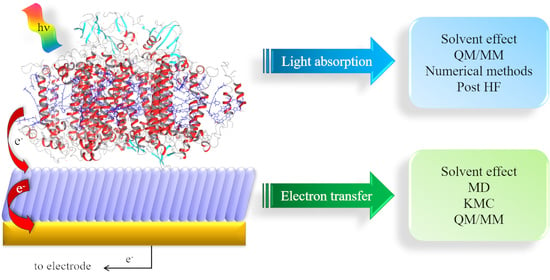
Artificial Photosynthesis: Is Computation Ready for the Challenge Ahead?
Abstract
:1. Introduction
2. Light Harvesting
2.1. QM/MM-Based Approaches
2.2. Numerical Modeling
2.3. Post HF Methods
2.4. Triplet–Triplet Energy Transfer
3. Protein ET Mechanisms
4. Protein/Surface Interactions
4.1. Protein–(Self-Assembly Monolayer (SAM))–Metal Interface
4.2. Adsorption on Low Dimensional Materials
5. Conclusions and Perspectives
Funding
Conflicts of Interest
References
- Cabuzel, T. 2030 Climate Target Plan. Available online: https://ec.europa.eu/clima/policies/eu-climate-action/2030_ctp_en (accessed on 30 November 2020).
- Walter, M.G.; Warren, E.L.; McKone, J.R.; Boettcher, S.W.; Mi, Q.; Santori, E.A.; Lewis, N.S. Solar Water Splitting Cells. Chem. Rev. 2010, 110, 6446–6473. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Lai, W.; Du, P. Catalytic Water Oxidation at Single Metal Sites. Energy Env. Sci. 2012, 5, 8134–8157. [Google Scholar] [CrossRef]
- Blankenship, R.E.; Tiede, D.M.; Barber, J.; Brudvig, G.W.; Fleming, G.; Ghirardi, M.; Gunner, M.R.; Junge, W.; Kramer, D.M.; Melis, A.; et al. Comparing Photosynthetic and Photovoltaic Efficiencies and Recognizing the Potential for Improvement. Science 2011, 332, 805–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janna Olmos, J.D.; Kargul, J. A Quest for the Artificial Leaf. Int. J. Biochem. Cell Biol. 2015, 66, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Osella, S.; Kargul, J.; Izzo, M.; Trzaskowski, B. Architecture and Function of Biohybrid Solar Cell and Solar-to-Fuel Nanodevices. In Theory and Simulation in Physics for Materials Applications: Cutting-Edge Techniques in Theoretical and Computational Materials Science; Levchenko, E.V., Dappe, Y.J., Ori, G., Eds.; Springer Series in Materials Science; Springer International Publishing: Cham, Switzerland, 2020; pp. 227–274. [Google Scholar] [CrossRef]
- Kamran, M.; Delgado, J.D.; Friebe, V.; Aartsma, T.J.; Frese, R.N. Photosynthetic Protein Complexes as Bio-Photovoltaic Building Blocks Retaining a High Internal Quantum Efficiency. Biomacromolecules 2014, 15, 2833–2838. [Google Scholar] [CrossRef]
- Kiliszek, M.; Harputlu, E.; Szalkowski, M.; Kowalska, D.; Unlu, C.G.; Haniewicz, P.; Abram, M.; Wiwatowski, K.; Niedziółka-Jönsson, J.; Maćkowski, S.; et al. Orientation of Photosystem I on Graphene through Cytochrome C553 Leads to Improvement in Photocurrent Generation. J. Mater. Chem. A 2018, 6, 18615–18626. [Google Scholar] [CrossRef]
- Ocakoglu, K.; Krupnik, T.; van den Bosch, B.; Harputlu, E.; Gullo, M.P.; Olmos, J.D.J.; Yildirimcan, S.; Gupta, R.K.; Yakuphanoglu, F.; Barbieri, A.; et al. Photosystem I-Based Biophotovoltaics on Nanostructured Hematite. Adv. Funct. Mater. 2014, 24, 7467–7477. [Google Scholar] [CrossRef]
- Amunts, A.; Drory, O.; Nelson, N. The Structure of a Plant Photosystem I Supercomplex at 3.4 A Resolution. Nature 2007, 447, 58–63. [Google Scholar] [CrossRef]
- Umena, Y.; Kawakami, K.; Shen, J.-R.; Kamiya, N. Crystal Structure of Oxygen-Evolving Photosystem II at a Resolution of 1.9 Å. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef]
- Mirkovic, T.; Ostroumov, E.E.; Anna, J.M.; van Grondelle, R.G.; Scholes, G.D. Light Absorption and Energy Transfer in the Antenna Complexes of Photosynthetic Organisms. Chem. Rev. 2017, 117, 249–293. [Google Scholar] [CrossRef]
- Kargul, J.; Bubak, G.; Andryianau, G. Biophotovoltaic Systems Based on Photosynthetic Complexes. In Encyclopedia of Interfacial Chemistry: Surface Science and Electrochemistry; Wandelt, K., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 7, pp. 43–63. ISBN 9780128097397. [Google Scholar]
- Liguori, N.; Croce, R.; Marrink, S.J.; Thallmair, S. Molecular Dynamics Simulations in Photosynthesis. Photosynth Res 2020, 144, 273–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segatta, F.; Cupellini, L.; Garavelli, M.; Mennucci, B. Quantum Chemical Modeling of the Photoinduced Activity of Multichromophoric Biosystems: Focus Review. Chem. Rev. 2019, 119, 9361–9380. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.J.; Mennucci, B. Delocalized Excitons in Natural Light-Harvesting Complexes. Rev. Mod. Phys. 2018, 90, 035003. [Google Scholar] [CrossRef] [Green Version]
- Curutchet, C.; Mennucci, B. Quantum Chemical Studies of Light Harvesting. Chem. Rev. 2017, 117, 294–343. [Google Scholar] [CrossRef] [PubMed]
- Cupellini, L.; Corbella, M.; Mennucci, B.; Curutchet, C. Electronic Energy Transfer in Biomacromolecules. Wires Comput. Mol. Sci. 2019, 9, e1392. [Google Scholar] [CrossRef] [Green Version]
- Chenu, A.; Scholes, G.D. Coherence in Energy Transfer and Photosynthesis. Annu Rev Phys Chem 2015, 66, 69–96. [Google Scholar] [CrossRef] [Green Version]
- Croce, R.; van Amerongen, H. Natural Strategies for Photosynthetic Light Harvesting. Nat. Chem. Biol. 2014, 10, 492–501. [Google Scholar] [CrossRef]
- Anna, J.M.; Scholes, G.D.; van Grondelle, R. A Little Coherence in Photosynthetic Light Harvesting. BioScience 2014, 64, 14–25. [Google Scholar] [CrossRef] [Green Version]
- Renger, T.; Müh, F. Understanding Photosynthetic Light-Harvesting: A Bottom up Theoretical Approach. Phys. Chem. Chem. Phys. 2013, 15, 3348–3371. [Google Scholar] [CrossRef] [Green Version]
- Pachón, L.A.; Brumer, P. Computational Methodologies and Physical Insights into Electronic Energy Transfer in Photosynthetic Light-Harvesting Complexes. Phys. Chem. Chem. Phys. 2012, 14, 10094–10108. [Google Scholar] [CrossRef] [Green Version]
- Fleming, G.R.; Schlau-Cohen, G.S.; Amarnath, K.; Zaks, J. Design Principles of Photosynthetic Light-Harvesting. Faraday Discuss. 2012, 155, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Scholes, G.D.; Mirkovic, T.; Turner, D.B.; Fassioli, F.; Buchleitner, A. Solar Light Harvesting by Energy Transfer: From Ecology to Coherence. Energy Env. Sci. 2012, 5, 9374–9393. [Google Scholar] [CrossRef]
- Subotnik, J.E.; Alguire, E.C.; Ou, Q.; Landry, B.R.; Fatehi, S. The Requisite Electronic Structure Theory To Describe Photoexcited Nonadiabatic Dynamics: Nonadiabatic Derivative Couplings and Diabatic Electronic Couplings. Acc. Chem. Res. 2015, 48, 1340–1350. [Google Scholar] [CrossRef]
- Persico, M.; Granucci, G. An Overview of Nonadiabatic Dynamics Simulations Methods, with Focus on the Direct Approach versus the Fitting of Potential Energy Surfaces. Theor. Chem Acc. 2014, 133, 1526. [Google Scholar] [CrossRef]
- Tavernelli, I. Nonadiabatic Molecular Dynamics Simulations: Synergies between Theory and Experiments. Acc. Chem. Res. 2015, 48, 792–800. [Google Scholar] [CrossRef]
- Barbatti, M. Nonadiabatic Dynamics with Trajectory Surface Hopping Method. Wires Comput. Mol. Sci. 2011, 1, 620–633. [Google Scholar] [CrossRef]
- Tully, J.C. Perspective: Nonadiabatic Dynamics Theory. J. Chem. Phys. 2012, 137, 22A301. [Google Scholar] [CrossRef]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum Mechanical Continuum Solvation Models. Chem. Rev. 2005, 105, 2999–3094. [Google Scholar] [CrossRef]
- Senn, H.M.; Thiel, W. QM/MM Methods for Biomolecular Systems. Angew. Chem. Int. Ed. 2009, 48, 1198–1229. [Google Scholar] [CrossRef]
- Frushicheva, M.P.; Mills, M.J.; Schopf, P.; Singh, M.K.; Prasad, R.B.; Warshel, A. Computer Aided Enzyme Design and Catalytic Concepts. Curr. Opin. Chem. Biol. 2014, 21, 56–62. [Google Scholar] [CrossRef] [Green Version]
- Boulanger, E.; Thiel, W. Solvent Boundary Potentials for Hybrid QM/MM Computations Using Classical Drude Oscillators: A Fully Polarizable Model. J. Chem. Theory Comput. 2012, 8, 4527–4538. [Google Scholar] [CrossRef] [PubMed]
- Frenkel, J. On the Transformation of light into Heat in Solids. Phys. Rev. 1931, 37, 17. [Google Scholar] [CrossRef]
- Iozzi, M.F.; Mennucci, B.; Tomasi, J.; Cammi, R. Excitation Energy Transfer (EET) between Molecules in Condensed Matter: A Novel Application of the Polarizable Continuum Model (PCM). J Chem Phys 2004, 120, 7029–7040. [Google Scholar] [CrossRef] [PubMed]
- Krueger, B.P.; Scholes, G.D.; Fleming, G.R. Calculation of Couplings and Energy-Transfer Pathways between the Pigments of LH2 by the Ab Initio Transition Density Cube Method. J. Phys. Chem. B 1998, 102, 5378–5386. [Google Scholar] [CrossRef]
- Caricato, M.; Curutchet, C.; Mennucci, B.; Scalmani, G. Electronic Couplings for Resonance Energy Transfer from CCSD Calculations: From Isolated to Solvated Systems. J. Chem. Theory Comput. 2015, 11, 5219–5228. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Losa, A.; Curutchet, C.; Galván, I.F.; Mennucci, B. Quantum Mechanical Methods Applied to Excitation Energy Transfer: A Comparative Analysis on Excitation Energies and Electronic Couplings. J. Chem. Phys. 2008, 129, 034104. [Google Scholar] [CrossRef]
- Fückel, B.; Köhn, A.; Harding, M.E.; Diezemann, G.; Hinze, G.; Basché, T.; Gauss, J. Theoretical Investigation of Electronic Excitation Energy Transfer in Bichromophoric Assemblies. J. Chem. Phys. 2008, 128, 074505. [Google Scholar] [CrossRef]
- Rebentrost, P.; Mohseni, M.; Kassal, I.; Lloyd, S.; Aspuru-Guzik, A. Environment-Assisted Quantum Transport. New J. Phys. 2009, 11, 033003. [Google Scholar] [CrossRef]
- Plenio, M.B.; Huelga, S.F. Dephasing-Assisted Transport: Quantum Networks and Biomolecules. New J. Phys. 2008, 10, 113019. [Google Scholar] [CrossRef]
- Novoderezhkin, V.I.; van Grondelle, R. Physical Origins and Models of Energy Transfer in Photosynthetic Light-Harvesting. Phys. Chem. Chem. Phys. 2010, 12, 7352–7365. [Google Scholar] [CrossRef]
- Novoderezhkin, V.; van Grondelle, R. Spectra and Dynamics in the B800 Antenna: Comparing Hierarchical Equations, Redfield and Förster Theories. J. Phys. Chem. B 2013, 117, 11076–11090. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, A.; Fleming, G.R. Unified Treatment of Quantum Coherent and Incoherent Hopping Dynamics in Electronic Energy Transfer: Reduced Hierarchy Equation Approach. J. Chem. Phys. 2009, 130, 234111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Q.; Chen, L.; Nan, G.; Xu, R.-X.; Yan, Y. Efficient Hierarchical Liouville Space Propagator to Quantum Dissipative Dynamics. J. Chem. Phys. 2009, 130, 084105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, C.; Neugebauer, J. First-Principles Calculation of Electronic Spectra of Light-Harvesting Complex II. Phys. Chem. Chem. Phys. 2011, 13, 10475–10490. [Google Scholar] [CrossRef] [PubMed]
- Casida, M.E.; Wesołowski, T.A. Generalization of the Kohn–Sham Equations with Constrained Electron Density Formalism and Its Time-Dependent Response Theory Formulation. Int. J. Quantum Chem. 2004, 96, 577–588. [Google Scholar] [CrossRef]
- Wesolowski, T.A.; Warshel, A. Frozen Density Functional Approach for Ab Initio Calculations of Solvated Molecules. J. Phys. Chem. 1993, 97, 8050–8053. [Google Scholar] [CrossRef]
- Aksu, H.; Schubert, A.; Geva, E.; Dunietz, B.D. Explaining Spectral Asymmetries and Excitonic Characters of the Core Pigment Pairs in the Bacterial Reaction Center Using a Screened Range-Separated Hybrid Functional. J. Phys. Chem. B 2019, 123, 8970–8975. [Google Scholar] [CrossRef]
- Bhandari, S.; Cheung, M.S.; Geva, E.; Kronik, L.; Dunietz, B.D. Fundamental Gaps of Condensed-Phase Organic Semiconductors from Single-Molecule Calculations Using Polarization-Consistent Optimally Tuned Screened Range-Separated Hybrid Functionals. J. Chem. Theory Comput. 2018, 14, 6287–6294. [Google Scholar] [CrossRef]
- Casida, M.E. Time-Dependent Density Functional Response Theory for Molecules. In Recent Advances in Density Functional Methods; Recent Advances in Computational Chemistry; World Scientific: Singapore, 1995; Volume 1, pp. 155–192. [Google Scholar] [CrossRef]
- Jornet-Somoza, J.; Lebedeva, I. Real-Time Propagation TDDFT and Density Analysis for Exciton Coupling Calculations in Large Systems. J. Chem. Theory Comput. 2019, 15, 3743–3754. [Google Scholar] [CrossRef] [Green Version]
- Lopata, K.; Govind, N. Modeling Fast Electron Dynamics with Real-Time Time-Dependent Density Functional Theory: Application to Small Molecules and Chromophores. J. Chem. Theory Comput. 2011, 7, 1344–1355. [Google Scholar] [CrossRef]
- Takimoto, Y.; Vila, F.D.; Rehr, J.J. Real-Time Time-Dependent Density Functional Theory Approach for Frequency-Dependent Nonlinear Optical Response in Photonic Molecules. J. Chem. Phys. 2007, 127, 154114. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.S.; Voorhis, T.V. Dynamic Current Suppression and Gate Voltage Response in Metal−Molecule−Metal Junctions. Nano Lett. 2009, 9, 2671–2675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perdew, J.P.; Levy, M. Physical Content of the Exact Kohn-Sham Orbital Energies: Band Gaps and Derivative Discontinuities. Phys. Rev. Lett. 1983, 51, 1884–1887. [Google Scholar] [CrossRef]
- Kümmel, S.; Kronik, L. Orbital-Dependent Density Functionals: Theory and Applications. Rev. Mod. Phys. 2008, 80, 3–60. [Google Scholar] [CrossRef] [Green Version]
- Seidl, A.; Görling, A.; Vogl, P.; Majewski, J.A.; Levy, M. Generalized Kohn-Sham Schemes and the Band-Gap Problem. Phys. Rev. B 1996, 53, 3764–3774. [Google Scholar] [CrossRef]
- Sham, L.J.; Schlüter, M. Density-Functional Theory of the Energy Gap. Phys. Rev. Lett. 1983, 51, 1888–1891. [Google Scholar] [CrossRef]
- Mori-Sánchez, P.; Cohen, A.J.; Yang, W. Localization and Delocalization Errors in Density Functional Theory and Implications for Band-Gap Prediction. Phys. Rev. Lett. 2008, 100, 146401. [Google Scholar] [CrossRef] [Green Version]
- Segatta, F.; Cupellini, L.; Jurinovich, S.; Mukamel, S.; Dapor, M.; Taioli, S.; Garavelli, M.; Mennucci, B. A Quantum Chemical Interpretation of Two-Dimensional Electronic Spectroscopy of Light-Harvesting Complexes. J. Am. Chem. Soc. 2017, 139, 7558–7567. [Google Scholar] [CrossRef]
- Malmqvist, P.Å.; Pierloot, K.; Shahi, A.R.M.; Cramer, C.J.; Gagliardi, L. The Restricted Active Space Followed by Second-Order Perturbation Theory Method: Theory and Application to the Study of CuO2 and Cu2O2 Systems. J. Chem. Phys. 2008, 128, 204109. [Google Scholar] [CrossRef]
- Roos, B.O. The Complete Active Space Self-Consistent Field Method and Its Applications in Electronic Structure Calculations. In Advances in Chemical Physics; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1987; pp. 399–445. [Google Scholar] [CrossRef]
- Zhang, W.M.; Meier, T.; Chernyak, V.; Mukamel, S. Exciton-Migration and Three-Pulse Femtosecond Optical Spectroscopies of Photosynthetic Antenna Complexes. J. Chem. Phys. 1998, 108, 7763–7774. [Google Scholar] [CrossRef] [Green Version]
- Mukamel, S.; Abramavicius, D. Many-Body Approaches for Simulating Coherent Nonlinear Spectroscopies of Electronic and Vibrational Excitons. Chem. Rev. 2004, 104, 2073–2098. [Google Scholar] [CrossRef] [PubMed]
- Abramavicius, D.; Palmieri, B.; Voronine, D.V.; Šanda, F.; Mukamel, S. Coherent Multidimensional Optical Spectroscopy of Excitons in Molecular Aggregates; Quasiparticle versus Supermolecule Perspectives. Chem. Rev. 2009, 109, 2350–2408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cupellini, L.; Bondanza, M.; Nottoli, M.; Mennucci, B. Successes & Challenges in the Atomistic Modeling of Light-Harvesting and Its Photoregulation. Biochim. Et Biophys. Acta (Bba) Bioenerg. 2020, 1861, 148049. [Google Scholar] [CrossRef] [Green Version]
- Curutchet, C.; Muñoz-Losa, A.; Monti, S.; Kongsted, J.; Scholes, G.D.; Mennucci, B. Electronic Energy Transfer in Condensed Phase Studied by a Polarizable QM/MM Model. J. Chem. Theory Comput. 2009, 5, 1838–1848. [Google Scholar] [CrossRef] [PubMed]
- König, C.; Neugebauer, J. Protein Effects on the Optical Spectrum of the Fenna–Matthews–Olson Complex from Fully Quantum Chemical Calculations. J. Chem. Theory Comput. 2013, 9, 1808–1820. [Google Scholar] [CrossRef] [PubMed]
- Wesolowski, T.A.; Shedge, S.; Zhou, X. Frozen-Density Embedding Strategy for Multilevel Simulations of Electronic Structure. Chem. Rev. 2015, 115, 5891–5928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.-P.; Fleming, G.R.; Head-Gordon, M.; Head-Gordon, T. Excitation Energy Transfer in Condensed Media. J. Chem. Phys. 2001, 114, 3065–3072. [Google Scholar] [CrossRef] [Green Version]
- Scholes, G.D.; Curutchet, C.; Mennucci, B.; Cammi, R.; Tomasi, J. How Solvent Controls Electronic Energy Transfer and Light Harvesting. J. Phys. Chem. B 2007, 111, 6978–6982. [Google Scholar] [CrossRef]
- Kim, Y.; Morozov, D.; Stadnytskyi, V.; Savikhin, S.; Slipchenko, L.V. Predictive First-Principles Modeling of a Photosynthetic Antenna Protein: The Fenna–Matthews–Olson Complex. J. Phys. Chem. Lett. 2020, 11, 1636–1643. [Google Scholar] [CrossRef]
- DeFusco, A.; Minezawa, N.; Slipchenko, L.V.; Zahariev, F.; Gordon, M.S. Modeling Solvent Effects on Electronic Excited States. J. Phys. Chem. Lett. 2011, 2, 2184–2192. [Google Scholar] [CrossRef] [Green Version]
- Gordon, M.S.; Smith, Q.A.; Xu, P.; Slipchenko, L.V. Accurate First Principles Model Potentials for Intermolecular Interactions. Annu. Rev. Phys. Chem. 2013, 64, 553–578. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, P.K.; Acharya, A.; Ghosh, D.; Kosenkov, D.; Kaliman, I.; Shao, Y.; Krylov, A.I.; Slipchenko, L.V. Extension of the Effective Fragment Potential Method to Macromolecules. J. Phys. Chem. B 2016, 120, 6562–6574. [Google Scholar] [CrossRef] [PubMed]
- Kaliman, I.A.; Slipchenko, L.V. Hybrid MPI/OpenMP Parallelization of the Effective Fragment Potential Method in the Libefp Software Library. J. Comput. Chem. 2015, 36, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Kosenkov, D.; Vanovschi, V.; Williams, C.F.; Herbert, J.M.; Gordon, M.S.; Schmidt, M.W.; Slipchenko, L.V.; Krylov, A.I. Noncovalent Interactions in Extended Systems Described by the Effective Fragment Potential Method: Theory and Application to Nucleobase Oligomers. J. Phys. Chem. A 2010, 114, 12739–12754. [Google Scholar] [CrossRef] [Green Version]
- Gordon, M.S.; Fedorov, D.G.; Pruitt, S.R.; Slipchenko, L.V. Fragmentation Methods: A Route to Accurate Calculations on Large Systems. Chem. Rev. 2012, 112, 632–672. [Google Scholar] [CrossRef] [Green Version]
- Slipchenko, L.V. Solvation of the Excited States of Chromophores in Polarizable Environment: Orbital Relaxation versus Polarization. J. Phys. Chem. A 2010, 114, 8824–8830. [Google Scholar] [CrossRef]
- Slipchenko, L.V.; Gurunathan, P.K. Effective Fragment Potential Method: Past, Present, and Future. In Fragmentation; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 183–208. [Google Scholar] [CrossRef]
- Baker, L.A.; Habershon, S. Photosynthesis, Pigment–Protein Complexes and Electronic Energy Transport: Simple Models for Complicated Processes. Sci. Prog. 2017, 100, 313–330. [Google Scholar] [CrossRef]
- Yang, M.; Damjanović, A.; Vaswani, H.M.; Fleming, G.R. Energy Transfer in Photosystem I of Cyanobacteria Synechococcus Elongatus: Model Study with Structure-Based Semi-Empirical Hamiltonian and Experimental Spectral Density. Biophys. J. 2003, 85, 140–158. [Google Scholar] [CrossRef] [Green Version]
- Baker, L.A.; Habershon, S. Robustness, Efficiency, and Optimality in the Fenna-Matthews-Olson Photosynthetic Pigment-Protein Complex. J. Chem. Phys. 2015, 143, 105101. [Google Scholar] [CrossRef] [Green Version]
- Richter, M.; Fingerhut, B.P. Coarse-Grained Representation of the Quasi Adiabatic Propagator Path Integral for the Treatment of Non-Markovian Long-Time Bath Memory. J. Chem. Phys. 2017, 146, 214101. [Google Scholar] [CrossRef]
- Richter, M.; Fingerhut, B.P. Coupled Excitation Energy and Charge Transfer Dynamics in Reaction Centre Inspired Model Systems. Faraday Discuss. 2019, 216, 72–93. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S.; Waletzke, M. A Combination of Kohn–Sham Density Functional Theory and Multi-Reference Configuration Interaction Methods. J. Chem. Phys. 1999, 111, 5645–5655. [Google Scholar] [CrossRef]
- Knecht, S.; Marian, C.M.; Kongsted, J.; Mennucci, B. On the Photophysics of Carotenoids: A Multireference DFT Study of Peridinin. J. Phys. Chem. B 2013, 117, 13808–13815. [Google Scholar] [CrossRef] [PubMed]
- List, N.H.; Curutchet, C.; Knecht, S.; Mennucci, B.; Kongsted, J. Toward Reliable Prediction of the Energy Ladder in Multichromophoric Systems: A Benchmark Study on the FMO Light-Harvesting Complex. J. Chem. Theory Comput. 2013, 9, 4928–4938. [Google Scholar] [CrossRef] [PubMed]
- Vico, L.D.; Anda, A.; Osipov, V.A.; Madsen, A.Ø.; Hansen, T. Macrocycle Ring Deformation as the Secondary Design Principle for Light-Harvesting Complexes. PNAS 2018, 115, E9051–E9057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Send, R.; Kaila, V.R.I.; Sundholm, D. Reduction of the Virtual Space for Coupled-Cluster Excitation Energies of Large Molecules and Embedded Systems. J. Chem. Phys. 2011, 134, 214114. [Google Scholar] [CrossRef] [Green Version]
- Send, R.; Suomivuori, C.-M.; Kaila, V.R.I.; Sundholm, D. Coupled-Cluster Studies of Extensive Green Fluorescent Protein Models Using the Reduced Virtual Space Approach. J. Phys. Chem. B 2015, 119, 2933–2945. [Google Scholar] [CrossRef]
- Trofimov, A.B.; Schirmer, J. An Efficient Polarization Propagator Approach to Valence Electron Excitation Spectra. J. Phys. B At. Mol. Opt. Phys. 1995, 28, 2299–2324. [Google Scholar] [CrossRef]
- Schirmer, J. Beyond the Random-Phase Approximation: A New Approximation Scheme for the Polarization Propagator. Phys. Rev. A 1982, 26, 2395–2416. [Google Scholar] [CrossRef]
- Suomivuori, C.-M.; Fliegl, H.; Starikov, E.B.; Balaban, T.S.; Kaila, V.R.I.; Sundholm, D. Absorption Shifts of Diastereotopically Ligated Chlorophyll Dimers of Photosystem I. Phys. Chem. Chem. Phys. 2019, 21, 6851–6858. [Google Scholar] [CrossRef] [Green Version]
- Leng, X.; Jin, F.; Wei, M.; Ma, H.; Feng, J.; Ma, Y. Electronic Energy Transfer Studied by Many-Body Green’s Function Theory. J. Chem. Phys. 2019, 150, 164107. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Lastra, J.M.; Thygesen, K.S. Renormalization of Optical Excitations in Molecules near a Metal Surface. Phys. Rev. Lett. 2011, 106, 187402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duchemin, I.; Deutsch, T.; Blase, X. Short-Range to Long-Range Charge-Transfer Excitations in the Zincbacteriochlorin-Bacteriochlorin Complex: A Bethe-Salpeter Study. Phys. Rev. Lett. 2012, 109, 167801. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Ma, Y.; Mu, J.; Liu, C.; Rohlfing, M. Charge-Transfer Excited States in Aqueous DNA: Insights from Many-Body Green’s Function Theory. Phys. Rev. Lett. 2014, 112, 228301. [Google Scholar] [CrossRef] [PubMed]
- Ort, D.R.; Yocum, C.F. Oxygenic Photosynthesis: The Light Reactions; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1996. [Google Scholar]
- You, Z.-Q.; Hsu, C.-P. Ab Inito Study on Triplet Excitation Energy Transfer in Photosynthetic Light-Harvesting Complexes. J. Phys. Chem. A 2011, 115, 4092–4100. [Google Scholar] [CrossRef]
- Curutchet, C.; Voityuk, A.A. Distance Dependence of Triplet Energy Transfer in Water and Organic Solvents: A QM/MD Study. J. Phys. Chem. C 2012, 116, 22179–22185. [Google Scholar] [CrossRef]
- Mani, T.; Niedzwiedzki, D.M.; Vinogradov, S.A. Generation of Phosphorescent Triplet States via Photoinduced Electron Transfer: Energy and Electron Transfer Dynamics in Pt Porphyrin–Rhodamine B Dyads. J. Phys. Chem. A 2012, 116, 3598–3610. [Google Scholar] [CrossRef] [Green Version]
- Dexter, D.L. A Theory of Sensitized Luminescence in Solids. J. Chem. Phys. 1953, 21, 836–850. [Google Scholar] [CrossRef]
- Marcus, R.A. Chemical and Electrochemical Electron-Transfer Theory. Annu. Rev. Phys. Chem. 1964, 15, 155. [Google Scholar] [CrossRef]
- Closs, G.L.; Johnson, M.D.; Miller, J.R.; Piotrowiak, P. A Connection between Intramolecular Long-Range Electron, Hole, and Triplet Energy Transfers. J. Am. Chem. Soc. 1989, 111, 3751–3753. [Google Scholar] [CrossRef]
- Albinsson, B.; Mårtensson, J. Excitation Energy Transfer in Donor–Bridge–Acceptor Systems. Phys. Chem. Chem. Phys. 2010, 12, 7338–7351. [Google Scholar] [CrossRef] [PubMed]
- Vura-Weis, J.; Abdelwahed, S.H.; Shukla, R.; Rathore, R.; Ratner, M.A.; Wasielewski, M.R. Crossover from Single-Step Tunneling to Multistep Hopping for Molecular Triplet Energy Transfer. Science 2010, 328, 1547–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConnell, H.M. Intramolecular Charge Transfer in Aromatic Free Radicals. J. Chem. Phys. 1961, 35, 508–515. [Google Scholar] [CrossRef]
- Angerhofer, A.; Bornhäuser, F.; Gall, A.; Cogdell, R.J. Optical and Optically Detected Magnetic Resonance Investigation on Purple Photosynthetic Bacterial Antenna Complexes. Chem. Phys. 1995, 194, 259–274. [Google Scholar] [CrossRef]
- Gall, A.; Berera, R.; Alexandre, M.T.A.; Pascal, A.A.; Bordes, L.; Mendes-Pinto, M.M.; Andrianambinintsoa, S.; Stoitchkova, K.V.; Marin, A.; Valkunas, L.; et al. Molecular Adaptation of Photoprotection: Triplet States in Light-Harvesting Proteins. Biophys. J. 2011, 101, 934–942. [Google Scholar] [CrossRef] [Green Version]
- Peterman, E.J.; Dukker, F.M.; van Grondelle, R.; van Amerongen, H. Chlorophyll a and Carotenoid Triplet States in Light-Harvesting Complex II of Higher Plants. Biophys. J. 1995, 69, 2670–2678. [Google Scholar] [CrossRef] [Green Version]
- Ho, J.; Kish, E.; Méndez-Hernández, D.D.; WongCarter, K.; Pillai, S.; Kodis, G.; Niklas, J.; Poluektov, O.G.; Gust, D.; Moore, T.A.; et al. Triplet–Triplet Energy Transfer in Artificial and Natural Photosynthetic Antennas. PNAS 2017, 114, E5513–E5521. [Google Scholar] [CrossRef] [Green Version]
- Di Valentin, M.; Tait, C.E.; Salvadori, E.; Orian, L.; Polimeno, A.; Carbonera, D. Evidence for Water-Mediated Triplet–Triplet Energy Transfer in the Photoprotective Site of the Peridinin–Chlorophyll a–Protein. Biochim. Et Biophys. Acta (Bba) Bioenerg. 2014, 1837, 85–97. [Google Scholar] [CrossRef]
- Davidson, V.L. Protein Control of True, Gated, and Coupled Electron Transfer Reactions. Acc. Chem. Res. 2008, 41, 730–738. [Google Scholar] [CrossRef] [Green Version]
- Paquete, C.M.; Louro, R.O. Unveiling the Details of Electron Transfer in Multicenter Redox Proteins. Acc. Chem. Res. 2014, 47, 56–65. [Google Scholar] [CrossRef]
- Beratan, D.N.; Onuchic, J.N.; Winkler, J.R.; Gray, H.B. Electron-Tunneling Pathways in Proteins. Science 1992, 258, 1740–1741. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yu, Y.; Tian, S.; Petrik, I.; Bhagi, A.; Lu, Y. Metalloproteins Containing Cytochrome, Iron–Sulfur, or Copper Redox Centers. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [CrossRef] [PubMed]
- Moser, C.C.; Keske, J.M.; Warncke, K.; Farid, R.S.; Dutton, P.L. Nature of Biological Electron Transfer. Nature 1992, 355, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Page, C.C.; Moser, C.C.; Chen, X.; Dutton, P.L. Natural Engineering Principles of Electron Tunnelling in Biological Oxidation–Reduction. Nature 1999, 402, 47–52. [Google Scholar] [CrossRef]
- Blumberger, J. Recent Advances in the Theory and Molecular Simulation of Biological Electron Transfer Reactions. Chem. Rev. 2015, 115, 11191–11238. [Google Scholar] [CrossRef] [Green Version]
- Williamson, H.R.; Dow, B.A.; Davidson, V.L. Mechanisms for Control of Biological Electron Transfer Reactions. Bioorganic Chem. 2014, 57, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Warren, J.J.; Ener, M.E.; Vlček, A.; Winkler, J.R.; Gray, H.B. Electron Hopping through Proteins. Coord. Chem. Rev. 2012, 256, 2478–2487. [Google Scholar] [CrossRef] [Green Version]
- Giese, B.; Graber, M.; Cordes, M. Electron Transfer in Peptides and Proteins. Curr. Opin. Chem. Biol. 2008, 12, 755–759. [Google Scholar] [CrossRef]
- Nakamura, K.; Go, N. Function and Molecular Evolution of Multicopper Blue Proteins. Cell. Mol. Life Sci. 2005, 62, 2050–2066. [Google Scholar] [CrossRef]
- Solomon, E.I.; Heppner, D.E.; Johnston, E.M.; Ginsbach, J.W.; Cirera, J.; Qayyum, M.; Kieber-Emmons, M.T.; Kjaergaard, C.H.; Hadt, R.G.; Tian, L. Copper Active Sites in Biology. Chem. Rev. 2014, 114, 3659–3853. [Google Scholar] [CrossRef] [Green Version]
- Battistuzzi, G.; Borsari, M.; Canters, G.W.; de Waal, E.; Loschi, L.; Warmerdam, G.; Sola, M. Enthalpic and Entropic Contributions to the Mutational Changes in the Reduction Potential of Azurin. Biochemistry 2001, 40, 6707–6712. [Google Scholar] [CrossRef] [PubMed]
- Pascher, T.; Karlsson, B.G.; Nordling, M.; Malmström, B.G.; Vänngård, T. Reduction Potentials and Their PH Dependence in Site-Directed-Mutant Forms of Azurin from Pseudomonas Aeruginosa. Eur. J. Biochem. 1993, 212, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Battistuzzi, G.; Borsari, M.; Di Rocco, G.; Leonardi, A.; Ranieri, A.; Sola, M. Electrostatic Effects on the Thermodynamics of Protonation of Reduced Plastocyanin. ChemBioChem 2005, 6, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.; Hoitink, C.W.G.; Nar, H.; Huber, R.; Messerschmidt, A.; Canters, G.W. X-Ray Analysis and Spectroscopic Characterization of M121Q Azurin: A Copper Site Model for Stellacyanin. J. Mol. Biol. 1993, 229, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Banfield, M.J.; Dennison, C. The Role of Hydrogen Bonding at the Active Site of a Cupredoxin: The Phe114Pro Azurin Variant. Biochemistry 2006, 45, 8812–8822. [Google Scholar] [CrossRef]
- Gray, H.B.; Malmström, B.G.; Williams, R.J.P. Copper Coordination in Blue Proteins. J. Biol. Inorg. Chem. 2000, 5, 551–559. [Google Scholar] [CrossRef]
- Wittung-Stafshede, P.; Hill, M.G.; Gomez, E.; Di Bilio, A.J.; Karlsson, B.G.; Leckner, J.; Winkler, J.R.; Gray, H.B.; Malmström, B.G. Reduction Potentials of Blue and Purple Copper Proteins in Their Unfolded States: A Closer Look at Rack-Induced Coordination. JBIC 1998, 3, 367–370. [Google Scholar] [CrossRef]
- Wei, C.; Lazim, R.; Zhang, D. Importance of Polarization Effect in the Study of Metalloproteins: Application of Polarized Protein Specific Charge Scheme in Predicting the Reduction Potential of Azurin. Proteins: Struct. Funct. Bioinform. 2014, 82, 2209–2219. [Google Scholar] [CrossRef]
- Botuyan, M.V.; Toy-Palmer, A.; Chung, J.; Blake II, R.C.; Beroza, P.; Case, D.A.; Dyson, H.J. NMR Solution Structure of Cu(I) Rusticyanin FromThiobacillus Ferrooxidans: Structural Basis for the Extreme Acid Stability and Redox Potential. J. Mol. Biol. 1996, 263, 752–767. [Google Scholar] [CrossRef]
- Li, H.; Webb, S.P.; Ivanic, J.; Jensen, J.H. Determinants of the Relative Reduction Potentials of Type-1 Copper Sites in Proteins. J. Am. Chem. Soc. 2004, 126, 8010–8019. [Google Scholar] [CrossRef]
- Hadt, R.G.; Sun, N.; Marshall, N.M.; Hodgson, K.O.; Hedman, B.; Lu, Y.; Solomon, E.I. Spectroscopic and DFT Studies of Second-Sphere Variants of the Type 1 Copper Site in Azurin: Covalent and Nonlocal Electrostatic Contributions to Reduction Potentials. J. Am. Chem. Soc. 2012, 134, 16701–16716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurd, C.A.; Besley, N.A.; Robinson, D. A QM/MM Study of the Nature of the Entatic State in Plastocyanin. J. Comput. Chem. 2017, 38, 1431–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, N.J.; Blanford, C.F.; Warwicker, J.; de Visser, S.P. Prediction of Reduction Potentials of Copper Proteins with Continuum Electrostatics and Density Functional Theory. Chem. A Eur. J. 2017, 23, 15436–15445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Cowley, R.E.; Tian, L.; Solomon, E.I. Mechanism of O2 Activation and Substrate Hydroxylation in Noncoupled Binuclear Copper Monooxygenases. PNAS 2016, 113, 12035–12040. [Google Scholar] [CrossRef] [Green Version]
- Bruschi, M.; Breglia, R.; Arrigoni, F.; Fantucci, P.; Gioia, L.D. Computational Approaches to the Prediction of the Redox Potentials of Iron and Copper Bioinorganic Systems. Int. J. Quantum Chem. 2016, 116, 1695–1705. [Google Scholar] [CrossRef] [Green Version]
- Warshel, A.; Papazyan, A.; Muegge, I. Microscopic and Semimacroscopic Redox Calculations: What Can and Cannot Be Learned from Continuum Models. JBIC 1997, 2, 143–152. [Google Scholar] [CrossRef]
- Shen, L.; Zeng, X.; Hu, H.; Hu, X.; Yang, W. Accurate Quantum Mechanical/Molecular Mechanical Calculations of Reduction Potentials in Azurin Variants. J. Chem. Theory Comput. 2018, 14, 4948–4957. [Google Scholar] [CrossRef]
- Hu, H.; Lu, Z.; Parks, J.M.; Burger, S.K.; Yang, W. Quantum Mechanics/Molecular Mechanics Minimum Free-Energy Path for Accurate Reaction Energetics in Solution and Enzymes: Sequential Sampling and Optimization on the Potential of Mean Force Surface. J. Chem. Phys. 2008, 128, 034105. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Lu, Z.; Yang, W. QM/MM Minimum Free-Energy Path: Methodology and Application to Triosephosphate Isomerase. J. Chem. Theory Comput. 2007, 3, 390–406. [Google Scholar] [CrossRef]
- Lin, X.; Hu, X.; Concepcion, J.J.; Chen, Z.; Liu, S.; Meyer, T.J.; Yang, W. Theoretical Study of Catalytic Mechanism for Single-Site Water Oxidation Process. PNAS 2012, 109, 15669–15672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, X.; Hu, H.; Hu, X.; Yang, W. Calculating Solution Redox Free Energies with Ab Initio Quantum Mechanical/Molecular Mechanical Minimum Free Energy Path Method. J. Chem. Phys. 2009, 130, 164111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prytkova, T.R.; Kurnikov, I.V.; Beratan, D.N. Coupling Coherence Distinguishes Structure Sensitivity in Protein Electron Transfer. Science 2007, 315, 622–625. [Google Scholar] [CrossRef] [Green Version]
- Winkler, J.R.; Gray, H.B. Long-Range Electron Tunneling. J. Am. Chem. Soc. 2014, 136, 2930–2939. [Google Scholar] [CrossRef] [PubMed]
- Moser, C.C.; Farid, T.A.; Chobot, S.E.; Dutton, P.L. Electron Tunneling Chains of Mitochondria. Biochim. Et Biophys. Acta (Bba) Bioenerg. 2006, 1757, 1096–1109. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, T.; Stuchebrukhov, A.A. Quantum Electron Tunneling in Respiratory Complex I. J. Phys. Chem. B 2011, 115, 5354–5364. [Google Scholar] [CrossRef] [Green Version]
- Heck, A.; Woiczikowski, P.B.; Kubař, T.; Giese, B.; Elstner, M.; Steinbrecher, T.B. Charge Transfer in Model Peptides: Obtaining Marcus Parameters from Molecular Simulation. J. Phys. Chem. B 2012, 116, 2284–2293. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.R.; Dinpajooh, M.; Matyushov, D.V. Polarizability of the Active Site in Enzymatic Catalysis: Cytochrome c. J. Phys. Chem. B 2019, 123, 10691–10699. [Google Scholar] [CrossRef]
- Martin, D.R.; Matyushov, D.V. Electron-Transfer Chain in Respiratory Complex I. Sci. Rep. 2017, 7, 5495. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.R.; Matyushov, D.V. Photosynthetic Diode: Electron Transport Rectification by Wetting the Quinone Cofactor. Phys. Chem. Chem. Phys. 2015, 17, 22523–22528. [Google Scholar] [CrossRef] [Green Version]
- Bortolotti, C.A.; Amadei, A.; Aschi, M.; Borsari, M.; Corni, S.; Sola, M.; Daidone, I. The Reversible Opening of Water Channels in Cytochrome c Modulates the Heme Iron Reduction Potential. J. Am. Chem. Soc. 2012, 134, 13670–13678. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.; Warshel, A. Capturing the Energetics of Water Insertion in Biological Systems: The Water Flooding Approach. Proteins: Struct. Funct. Bioinform. 2013, 81, 93–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leioatts, N.; Mertz, B.; Martínez-Mayorga, K.; Romo, T.D.; Pitman, M.C.; Feller, S.E.; Grossfield, A.; Brown, M.F. Retinal Ligand Mobility Explains Internal Hydration and Reconciles Active Rhodopsin Structures. Biochemistry 2014, 53, 376–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lasseter, T.L.; Clare, B.H.; Abbott, N.L.; Hamers, R.J. Covalently Modified Silicon and Diamond Surfaces: Resistance to Nonspecific Protein Adsorption and Optimization for Biosensing. J. Am. Chem. Soc. 2004, 126, 10220–10221. [Google Scholar] [CrossRef]
- Basset, C.; Harder, C.; Vidaud, C.; Déjugnat, C. Design of Double Stimuli-Responsive Polyelectrolyte Microcontainers for Protein Soft Encapsulation. Biomacromolecules 2010, 11, 806–814. [Google Scholar] [CrossRef]
- Messersmith, P.B. Multitasking in Tissues and Materials. Science 2008, 319, 1767–1768. [Google Scholar] [CrossRef]
- Sousa, S.R.; Moradas-Ferreira, P.; Saramago, B.; Viseu Melo, L.; Barbosa, M.A. Human Serum Albumin Adsorption on TiO2 from Single Protein Solutions and from Plasma. Langmuir 2004, 20, 9745–9754. [Google Scholar] [CrossRef]
- Wei, T.; Kaewtathip, S.; Shing, K. Buffer Effect on Protein Adsorption at Liquid/Solid Interface. J. Phys. Chem. C 2009, 113, 2053–2062. [Google Scholar] [CrossRef]
- Su, T.J.; Green, R.J.; Wang, Y.; Murphy, E.F.; Lu, J.R.; Ivkov, R.; Satija, S.K. Adsorption of Lysozyme onto the Silicon Oxide Surface Chemically Grafted with a Monolayer of Pentadecyl-1-Ol. Langmuir 2000, 16, 4999–5007. [Google Scholar] [CrossRef]
- Kim, D.T.; Blanch, H.W.; Radke, C.J. Direct Imaging of Lysozyme Adsorption onto Mica by Atomic Force Microscopy. Langmuir 2002, 18, 5841–5850. [Google Scholar] [CrossRef]
- Shaw, D.E.; Maragakis, P.; Lindorff-Larsen, K.; Piana, S.; Dror, R.O.; Eastwood, M.P.; Bank, J.A.; Jumper, J.M.; Salmon, J.K.; Shan, Y.; et al. Atomic-Level Characterization of the Structural Dynamics of Proteins. Science 2010, 330, 341–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Mu, S.; Nakano, A.; Shing, K. A Hybrid Multi-Loop Genetic-Algorithm/Simplex/Spatial-Grid Method for Locating the Optimum Orientation of an Adsorbed Protein on a Solid Surface. Comput. Phys. Commun. 2009, 180, 669–674. [Google Scholar] [CrossRef]
- Wei, T.; Carignano, M.A.; Szleifer, I. Molecular Dynamics Simulation of Lysozyme Adsorption/Desorption on Hydrophobic Surfaces. J. Phys. Chem. B 2012, 116, 10189–10194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bostick, C.D.; Mukhopadhyay, S.; Pecht, I.; Sheves, M.; Cahen, D.; Lederman, D. Protein Bioelectronics: A Review of What We Do and Do Not Know. Rep. Prog. Phys. 2018, 81, 026601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bixon, M.; Jortner, J. Electron Transfer—from Isolated Molecules to Biomolecules. In Advances in Chemical Physics; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2007; pp. 35–202. [Google Scholar] [CrossRef]
- Rasor, J.P.; Voss, E. Enzyme-Catalyzed Processes in Pharmaceutical Industry. Appl. Catal. A Gen. 2001, 221, 145–158. [Google Scholar] [CrossRef]
- Ozboyaci, M.; Kokh, D.B.; Corni, S.; Wade, R.C. Modeling and Simulation of Protein–Surface Interactions: Achievements and Challenges. Quart. Rev. Biophys. 2016, 49, e4. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Chu, P.K.; Ding, C. Surface Modification of Titanium, Titanium Alloys, and Related Materials for Biomedical Applications. Mater. Sci. Eng. R Rep. 2004, 47, 49–121. [Google Scholar] [CrossRef] [Green Version]
- Alessandrini, A.; Salerno, M.; Frabboni, S.; Facci, P. Single-Metalloprotein Wet Biotransistor. Appl. Phys. Lett. 2005, 86, 133902. [Google Scholar] [CrossRef]
- Walsh, T.R. Pathways to Structure–Property Relationships of Peptide–Materials Interfaces: Challenges in Predicting Molecular Structures. Acc. Chem. Res. 2017, 50, 1617–1624. [Google Scholar] [CrossRef] [Green Version]
- Love, J.C.; Estroff, L.A.; Kriebel, J.K.; Nuzzo, R.G.; Whitesides, G.M. Self-Assembled Monolayers of Thiolates on Metals as a Form of Nanotechnology. Chem. Rev. 2005, 105, 1103–1170. [Google Scholar] [CrossRef]
- Zhou, J.; Zheng, J.; Jiang, S. Molecular Simulation Studies of the Orientation and Conformation of Cytochrome c Adsorbed on Self-Assembled Monolayers. J. Phys. Chem. B 2004, 108, 17418–17424. [Google Scholar] [CrossRef]
- Bewley, K.D.; Ellis, K.E.; Firer-Sherwood, M.A.; Elliott, S.J. Multi-Heme Proteins: Nature’s Electronic Multi-Purpose Tool. Biochim. Et Biophys. Acta (Bba) Bioenerg. 2013, 1827, 938–948. [Google Scholar] [CrossRef] [Green Version]
- Mowat, C.G.; Chapman, S.K. Multi-Heme Cytochromes—New Structures, New Chemistry. Dalton Trans. 2005, 21, 3381–3389. [Google Scholar] [CrossRef] [Green Version]
- Breuer, M.; Rosso, K.M.; Blumberger, J.; Butt, J.N. Multi-Haem Cytochromes in Shewanella Oneidensis MR-1: Structures, Functions and Opportunities. J. R. Soc. Interface 2015, 12, 20141117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauß, A.; Koslowski, T. Storage, Transport, Release: Heme Versatility in Nitrite Reductase Electron Transfer Studied by Molecular Dynamics Simulations. Phys. Chem. Chem. Phys. 2015, 17, 4483–4491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmeier, A.; Murphy, B.J.; Armstrong, F.A. A Multi-Heme Flavoenzyme as a Solar Conversion Catalyst. J. Am. Chem. Soc. 2014, 136, 12876–12879. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Ma, H.; Nakano, A. Decaheme Cytochrome MtrF Adsorption and Electron Transfer on Gold Surface. J. Phys. Chem. Lett. 2016, 7, 929–936. [Google Scholar] [CrossRef]
- Paissoni, C.; Spiliotopoulos, D.; Musco, G.; Spitaleri, A. GMXPBSA 2.1: A GROMACS Tool to Perform MM/PBSA and Computational Alanine Scanning. Comput. Phys. Commun. 2015, 186, 105–107. [Google Scholar] [CrossRef]
- Byun, H.S.; Pirbadian, S.; Nakano, A.; Shi, L.; El-Naggar, M.Y. Kinetic Monte Carlo Simulations and Molecular Conductance Measurements of the Bacterial Decaheme Cytochrome MtrF. ChemElectroChem 2014, 1, 1932–1939. [Google Scholar] [CrossRef]
- Nakano, C.M.; Byun, H.S.; Ma, H.; Wei, T.; El-Naggar, M.Y. A Framework for Stochastic Simulations and Visualization of Biological Electron-Transfer Dynamics. Comput. Phys. Commun. 2015, 193, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, M.P.; Aragonès, A.C.; Camarero, N.; Vilhena, J.G.; Ortega, M.; Zotti, L.A.; Pérez, R.; Cuevas, J.C.; Gorostiza, P.; Díez-Pérez, I. Bioengineering a Single-Protein Junction. J. Am. Chem. Soc. 2017, 139, 15337–15346. [Google Scholar] [CrossRef]
- Ortega, M.; Vilhena, J.G.; Zotti, L.A.; Díez-Pérez, I.; Cuevas, J.C.; Pérez, R. Tuning Structure and Dynamics of Blue Copper Azurin Junctions via Single Amino-Acid Mutations. Biomolecules 2019, 9, 611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scida, K.; Stege, P.W.; Haby, G.; Messina, G.A.; García, C.D. Recent Applications of Carbon-Based Nanomaterials in Analytical Chemistry: Critical Review. Anal. Chim. Acta 2011, 691, 6–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Liang, X.-J. Nano-Carbons as Theranostics. Theranostics 2012, 2, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Knecht, M.R. Biomolecular Material Recognition in Two Dimensions: Peptide Binding to Graphene, h-BN, and MoS2 Nanosheets as Unique Bioconjugates. Bioconjugate Chem. 2019, 30, 2727–2750. [Google Scholar] [CrossRef]
- Hughes, Z.E.; Walsh, T.R. What Makes a Good Graphene-Binding Peptide? Adsorption of Amino Acids and Peptides at Aqueous Graphene Interfaces. J. Mater. Chem. B 2015, 3, 3211–3221. [Google Scholar] [CrossRef] [Green Version]
- Hughes, Z.E.; Walsh, T.R. Probing Nano-Patterned Peptide Self-Organisation at the Aqueous Graphene Interface. Nanoscale 2017, 10, 302–311. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.S.; Kuang, Z.; Ngo, Y.H.; Farmer, B.L.; Naik, R.R. Biotic–Abiotic Interactions: Factors That Influence Peptide–Graphene Interactions. Acs Appl. Mater. Interfaces 2015, 7, 20447–20453. [Google Scholar] [CrossRef]
- Heinz, H.; Farmer, B.L.; Pandey, R.B.; Slocik, J.M.; Patnaik, S.S.; Pachter, R.; Naik, R.R. Nature of Molecular Interactions of Peptides with Gold, Palladium, and Pd−Au Bimetal Surfaces in Aqueous Solution. J. Am. Chem. Soc. 2009, 131, 9704–9714. [Google Scholar] [CrossRef]
- Yu, J.; Becker, M.L.; Carri, G.A. The Influence of Amino Acid Sequence and Functionality on the Binding Process of Peptides onto Gold Surfaces. Langmuir 2012, 28, 1408–1417. [Google Scholar] [CrossRef]
- Penna, M.J.; Mijajlovic, M.; Biggs, M.J. Molecular-Level Understanding of Protein Adsorption at the Interface between Water and a Strongly Interacting Uncharged Solid Surface. J. Am. Chem. Soc. 2014, 136, 5323–5331. [Google Scholar] [CrossRef]
- Zhang, C.; Li, X.; Wang, Z.; Huang, X.; Ge, Z.; Hu, B. Influence of Structured Water Layers on Protein Adsorption Process: A Case Study of Cytochrome c and Carbon Nanotube Interactions and Its Implications. J. Phys. Chem. B 2020, 124, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Paggi, D.; Martín, D.F.; DeBiase, P.M.; Hildebrandt, P.; Martí, M.A.; Murgida, D.H. Molecular Basis of Coupled Protein and Electron Transfer Dynamics of Cytochrome c in Biomimetic Complexes. J. Am. Chem. Soc. 2010, 132, 5769–5778. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Kim, Y.-K.; Shin, D.; Ryoo, S.-R.; Hong, B.H.; Min, D.-H. Biomedical Applications of Graphene and Graphene Oxide. Acc. Chem. Res. 2013, 46, 2211–2224. [Google Scholar] [CrossRef] [PubMed]
- Patila, M.; Pavlidis, I.V.; Diamanti, E.K.; Katapodis, P.; Gournis, D.; Stamatis, H. Enhancement of Cytochrome c Catalytic Behaviour by Affecting the Heme Environment Using Functionalized Carbon-Based Nanomaterials. Process Biochem. 2013, 48, 1010–1017. [Google Scholar] [CrossRef]
- Shao, Q.; Wu, P.; Xu, X.; Zhang, H.; Cai, C. Insight into the Effects of Graphene Oxide Sheets on the Conformation and Activity of Glucose Oxidase: Towards Developing a Nanomaterial-Based Protein Conformation Assay. Phys. Chem. Chem. Phys. 2012, 14, 9076–9085. [Google Scholar] [CrossRef]
- Vincent, K.A.; Li, X.; Blanford, C.F.; Belsey, N.A.; Weiner, J.H.; Armstrong, F.A. Enzymatic Catalysis on Conducting Graphite Particles. Nat. Chem. Biol. 2007, 3, 761–762. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, C.; Ju, E.; Ren, J.; Qu, X. Contrasting Modulation of Enzyme Activity Exhibited by Graphene Oxide and Reduced Graphene. Chem. Commun. 2013, 49, 8611–8613. [Google Scholar] [CrossRef]
- Zhao, D.; Li, L.; Zhou, J. Simulation Insight into the Cytochrome c Adsorption on Graphene and Graphene Oxide Surfaces. Appl. Surf. Sci. 2018, 428, 825–834. [Google Scholar] [CrossRef]
- Kowalska, A.M.; Trzaskowski, B.; Osella, S. Assessing the Charge Transfer at the Cytochrome C553/Graphene Interface: A Multiscale Investigation. J. Phys. Chem. C 2018, 122, 29405–29413. [Google Scholar] [CrossRef] [Green Version]
- Achari, P.F.; Bejagam, K.K.; Singh, S.; Deshmukh, S.A. Development of Non-Bonded Interaction Parameters between Hexagonal Boron-Nitride and Water. Comput. Mater. Sci. 2019, 161, 339–345. [Google Scholar] [CrossRef]
- Govind Rajan, A.; Strano, M.S.; Blankschtein, D. Ab Initio Molecular Dynamics and Lattice Dynamics-Based Force Field for Modeling Hexagonal Boron Nitride in Mechanical and Interfacial Applications. J. Phys. Chem. Lett. 2018, 9, 1584–1591. [Google Scholar] [CrossRef] [PubMed]
- Mudedla, S.K.; Balamurugan, K.; Subramanian, V. Unravelling the Structural Changes in α-Helical Peptides on Interaction with Convex, Concave, and Planar Surfaces of Boron-Nitride-Based Nanomaterials. J. Phys. Chem. C 2016, 120, 28246–28260. [Google Scholar] [CrossRef]
- Xiao, M.; Wei, S.; Li, Y.; Jasensky, J.; Chen, J.; Brooks, C.L.; Chen, Z. Molecular Interactions between Single Layered MoS2 and Biological Molecules. Chem. Sci. 2018, 9, 1769–1773. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Li, M.; Liang, H.; Lou, H. Adsorption Patterns of Aromatic Amino Acids on Monolayer MoS2 and Au-Modified MoS2 Surfaces: A First-Principles Study. Comput. Theor. Chem. 2017, 1118, 115–122. [Google Scholar] [CrossRef]
- Nicolaï, A.; Barrios Pérez, M.D.; Delarue, P.; Meunier, V.; Drndić, M.; Senet, P. Molecular Dynamics Investigation of Polylysine Peptide Translocation through MoS2 Nanopores. J. Phys. Chem. B 2019, 123, 2342–2353. [Google Scholar] [CrossRef]
- Mudedla, S.K.; Murugan, N.A.; Agren, H. Free Energy Landscape for Alpha-Helix to Beta-Sheet Interconversion in Small Amyloid Forming Peptide under Nanoconfinement. J. Phys. Chem. B 2018, 122, 9654–9664. [Google Scholar] [CrossRef]
- Gu, Z.; Luna, P.D.; Yang, Z.; Zhou, R. Structural Influence of Proteins upon Adsorption to MoS2 Nanomaterials: Comparison of MoS2 Force Field Parameters. Phys. Chem. Chem. Phys. 2017, 19, 3039–3045. [Google Scholar] [CrossRef]














Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osella, S. Artificial Photosynthesis: Is Computation Ready for the Challenge Ahead? Nanomaterials 2021, 11, 299. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11020299
Osella S. Artificial Photosynthesis: Is Computation Ready for the Challenge Ahead? Nanomaterials. 2021; 11(2):299. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11020299
Chicago/Turabian StyleOsella, Silvio. 2021. "Artificial Photosynthesis: Is Computation Ready for the Challenge Ahead?" Nanomaterials 11, no. 2: 299. https://0-doi-org.brum.beds.ac.uk/10.3390/nano11020299