Real-Time Detection of Staphylococcus Aureus Using Whispering Gallery Mode Optical Microdisks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microdisk Fabrication
2.2. Plasmids and LysK Purification
2.3. Surface Functionalization
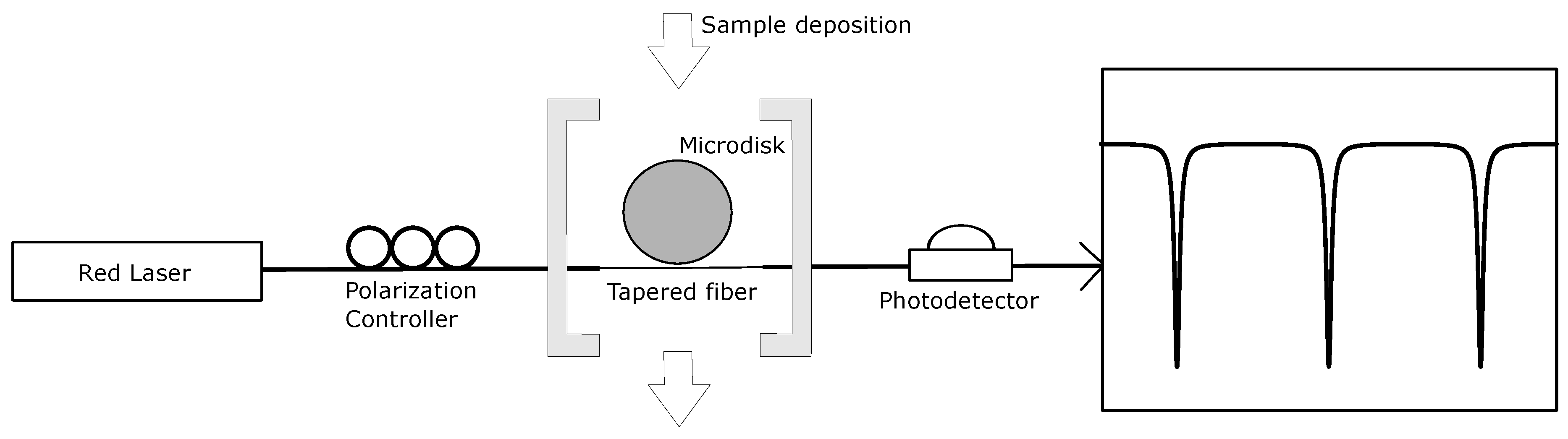
2.4. Optical Characterization Setup
2.5. Bacterial Strains and Maintenance
3. Results and Discussion
3.1. Fabrication and Characterization
3.2. Bacterial Binding
3.3. Specificity of LysK
3.4. Concentration-Dependent Shift
3.5. Bacterial Surface Density
3.6. Variability in Binding
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Thapa, B. Antimicrobial resistance: A global threat. Int. J. Infect. Microbiol. 2013, 1, 41–42. [Google Scholar] [CrossRef]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Klevens, R.M.; Morrison, M.A.; Nadle, J.; Petit, S.; Gershman, K.; Ray, S.; Harrison, L.H.; Lynfield, R.; Dumyati, G.; Townes, J.M.; et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. J. Am. Med. Assoc. 2007, 298, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Van Hal, S.; Stark, D.; Lockwood, B.; Marriott, D.; Harkness, J. Methicillin-resistant Staphylococcus aureus (MRSA) detection: Comparison of two molecular methods (IDI-MRSA PCR assay and GenoType MRSA Direct PCR assay) with three selective MRSA agars (MRSA ID, MRSASelect, and CHROMagar MRSA) for use with infection-control swabs. J. Clin. Microbiol. 2007, 45, 2486–2490. [Google Scholar] [PubMed]
- Luchansky, M.S.; Bailey, R.C. High-Q optical sensors for chemical and biological analysis. Anal. Chem. 2011, 84, 793–821. [Google Scholar] [CrossRef] [PubMed]
- Vahala, K.J. Optical microcavities. Nature 2003, 424, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Massad-Ivanir, N.; Shtenberg, G.; Tzur, A.; Krepker, M.A.; Segal, E. Engineering nanostructured porous SiO2 surfaces for bacteria detection via “direct cell capture”. Anal. Chem. 2011, 83, 3282–3289. [Google Scholar] [PubMed]
- Lopez-Yglesias, X.; Gamba, J.M.; Flagan, R.C. The physics of extreme sensitivity in whispering gallery mode optical biosensors. J. Appl. Phys. 2012, 111. [Google Scholar] [CrossRef]
- Foreman, M.R.; Swaim, J.D.; Vollmer, F. Whispering gallery mode sensors. Adv. Opt. Photonics 2015, 7, 168–240. [Google Scholar]
- Arnold, S.; Keng, D.; Shopova, S.; Holler, S.; Zurawsky, W.; Vollmer, F. Whispering gallery mode carousel—A photonic mechanism for enhanced nanoparticle detection in biosensing. Opt. Express 2009, 17, 6230–6238. [Google Scholar]
- Anderson, M.E.; O’Brien, E.C.; Grayek, E.N.; Hermansen, J.K.; Hunt, H.K. The Detection of Helicobacter hepaticus Using Whispering-Gallery Mode Microcavity Optical Sensors. Biosensors 2015, 5, 562–576. [Google Scholar]
- Becker, S.C.; Foster-Frey, J.; Donovan, D.M. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiol. Lett. 2008, 287, 185–191. [Google Scholar]
- Nelson, D.; Loomis, L.; Fischetti, V.A. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 2001, 98, 4107–4112. [Google Scholar]
- Chibli, H.; Ghali, H.; Park, S.; Peter, Y.A.; Nadeau, J.L. Immobilized phage proteins for specific detection of staphylococci. Analyst 2014, 139, 179–186. [Google Scholar] [PubMed]
- Bergeron, S.; Vanier, F.; Peter, Y.A. Silica Microdisk Coupled Resonator Optical Waveguide. In Proceedings of the 2009 IEEE/LEOS International Conference on Optical MEMS and Nanophotonics, Clearwater, FL, USA, 17–20 August 2009; pp. 73–74.
- Becker, S.C.; Foster-Frey, J.; Stodola, A.J.; Anacker, D.; Donovan, D.M. Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene 2009, 443, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Donovan, D.M.; Lardeo, M.; Foster-Frey, J. Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin. FEMS Microbiol. Lett. 2006, 265, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Ghali, H.; Bianucci, P.; Peter, Y.A. Wavelength Shift in a Whispering Gallery Microdisk due to Bacterial Sensing: A Theoretical Approach. J. Biomed. Optics 2016. submitted for publication. [Google Scholar]
- Guntupalli, R.; Sorokulova, I.; Olsen, E.; Globa, L.; Pustovyy, O.; Vodyanoy, V. Biosensor for detection of antibiotic resistant Staphylococcus bacteria. J. Vis. Exp. 2013. [Google Scholar] [CrossRef] [PubMed]




| No Dilution | Diluted 10× | Diluted 100× | Diluted 1000× |
|---|---|---|---|
| 0.22 ± 0.002 nm | 0.19 ± 0.002 nm | 0.15 ± 0.002 nm | 0.14 ± 0.002 nm |
| Parameter | Definition | Value | Unit |
|---|---|---|---|
| Excess polarizability | 4 × 2.12 × 10 | m | |
| Surface coverage | () | m | |
| Vacuum permittivity | 8.854 × 10 | F/m | |
| Relative permittivity of silica | 3.9 | ||
| k | Wavenumber () | 9.895 × 10 | nm |
| n | Refractive index of the microdisk | 1.457 | - |
| n | Refractive index of the buffer | 1.332 | - |
| h | Thickness of the microdisk | 800 | nm |
| R | Radius of the microdisk | 100 | m |
| m | Mass of one bacterium | 10 | g |
| l | Mode number () | 1442 | - |
| Bacterial Solution Concentration (CFU/mL) | Wavelength Shift (nm) | Surface Coverage () | Bacteria Bound to the Surface |
|---|---|---|---|
| 5 × 10 | 0.22 | 8.21 × | 31 |
| 5 × 10 | 0.19 | 7.15 × | 27 |
| 5 × 10 | 0.15 | 5.56 × | 21 |
| 5 × 10 | 0.14 | 5.3 × | 20 |
| Bacterial Solution Concentration (CFU/mL) | Wavelength Shift (nm) | Standard Deviation (nm) |
|---|---|---|
| 5 × | 0.22 | 0.01 |
| 5 × | 0.19 | 0.02 |
| 5 × | 0.15 | 0.04 |
| 5 × | 0.14 | 0.05 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghali, H.; Chibli, H.; Nadeau, J.L.; Bianucci, P.; Peter, Y.-A. Real-Time Detection of Staphylococcus Aureus Using Whispering Gallery Mode Optical Microdisks. Biosensors 2016, 6, 20. https://0-doi-org.brum.beds.ac.uk/10.3390/bios6020020
Ghali H, Chibli H, Nadeau JL, Bianucci P, Peter Y-A. Real-Time Detection of Staphylococcus Aureus Using Whispering Gallery Mode Optical Microdisks. Biosensors. 2016; 6(2):20. https://0-doi-org.brum.beds.ac.uk/10.3390/bios6020020
Chicago/Turabian StyleGhali, Hala, Hicham Chibli, Jay L. Nadeau, Pablo Bianucci, and Yves-Alain Peter. 2016. "Real-Time Detection of Staphylococcus Aureus Using Whispering Gallery Mode Optical Microdisks" Biosensors 6, no. 2: 20. https://0-doi-org.brum.beds.ac.uk/10.3390/bios6020020






