Label-Free Biosensor Detection of Endocrine Disrupting Compounds Using Engineered Estrogen Receptors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. ERαLBD Production and Purification
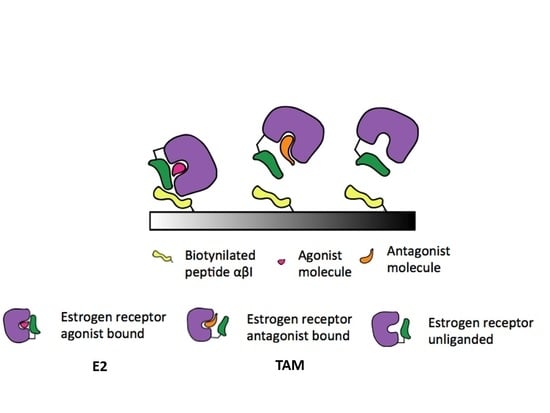
2.3. Design of the Binding Assay
2.4. SPR Measurements
2.4.1. ProteOn XPR36™
2.4.2. Biacore 3000
2.5. Water Sample Preparation by SPE
2.6. GC-MS Analysis
3. Results and Discussion
3.1. Optimisation of the Assay Parameters
3.2. Limit of Detection, Sensitivity and Specificity of the Assay for wtERαLBD and E2
3.3. Testing Water Samples
3.4. Array of ERα Receptors Mutants
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kortenkamp, A.; Martin, O.; Faust, M.; Evans, R.; McKinlay, R.F.; Orton, F.; Rosivatz, E. State of the art assessment of endocrine disrupters. Final Rep. 2011, 23, 1–135. [Google Scholar]
- Waring, R.H.; Harris, R.M. Endocrine disrupters: A human risk? Mol. Cell. Endocrinol. 2005, 244, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Jeselsohn, R.; Yelensky, R.; Buchwalter, G.; Frampton, G.; Meric-Bernstam, F.; Gonzalez-Angulo, A.M.; Ferrer-Lozano, J.; Perez-Fidalgo, J.A.; Cristofanilli, M.; Gomez, H.; et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin. Cancer Res. 2014, 20, 1757–1767. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.L.; Lester, J.N. Endocrine Disrupters in Drinking Water and Water Reuse; CRC Press: London, UK, 2002; pp. 219–266. [Google Scholar]
- Diamanti-Kandarakis, E.; Bourguignon, J.P.; Giudice, L.C.; Hauser, R.; Prins, G.S.; Soto, A.M.; Zoeller, R.T.; Gore, A.C. Endocrine-disrupting chemicals: An Endocrine Society scientific statement. Endocr. Rev. 2009, 30, 293–342. [Google Scholar] [CrossRef] [PubMed]
- Locatelli, M.; Sciascia, F.; Cifelli, R.; Malatesta, L.; Bruni, P.; Croce, F. Analytical methods for the endocrine disruptor compounds determination in environmental water samples. J. Chromatogr. A 2016, 1434, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.N.; Arukwe, A.; Ait-Aissa, S.; Bado-Nilles, A.; Balzamo, S.; Baun, A.; Belkin, S.; Blaha, L.; Brion, F.; Conti, D.; Creusot, N.; et al. Mixtures of chemical pollutants at European legislation safety concentrations: How safe are they? Toxicol. Sci. 2014, 141, 218–233. [Google Scholar] [CrossRef] [PubMed]
- Omar, T.F.T.; Ahmad, A.; Aris, A.Z.; Yusoff, F.M. Endocrine disrupting compounds (EDCs) in environmental matrices: Review of analytical strategies for pharmaceuticals, estrogenic hormones, and alkylphenol compounds. TrAC Trends Anal. Chem. 2016, 85, 241–259. [Google Scholar] [CrossRef]
- Marchesini, G.R.; Meulenberg, E.; Haasnoot, W.; Irth, H. Biosensor immunoassays for the detection of bisphenol A. Anal. Chim. Acta 2005, 528, 37–45. [Google Scholar] [CrossRef]
- Ojeda, I.; Lopez-Montero, J.; Moreno-Guzman, M.; Janegitz, B.C.; Gonzalez-Cortes, A.; Yanez-Sedeno, P.; Pingarron, J.M. Electrochemical immunosensor for rapid and sensitive determination of estradiol. Anal. Chim. Acta 2012, 743, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.M.; Dong, T. Ultrasensitive opto-microfluidic immunosensor integrating gold nanoparticle-enhanced chemiluminescence and highly stable organic photodetector. J. Biomed. Opt. 2014, 19, 30504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Du, B.; Li, H.; Xin, X.; Ma, H.; Wu, D.; Yan, L.; Wei, Q. Metal ions-based immunosensor for simultaneous determination of estradiol and diethylstilbestrol. Biosens. Bioelectron. 2014, 52, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Coille, I.; Reder, S.; Bucher, S.; Gauglitz, G. Comparison of two fluorescence immunoassay methods for the detection of endocrine disrupting chemicals in water. Biomol. Eng. 2002, 18, 273–280. [Google Scholar] [CrossRef]
- Beato, M. Gene regulation by steroid hormones. Cell 1989, 56, 335–344. [Google Scholar] [CrossRef]
- Katzenellenbogen, B.S.; Katzenellenbogen, J.A. Estrogen receptor transcription and transactivation: Estrogen receptor alpha and estrogen receptor beta: Regulation by selective estrogen receptor modulators and importance in breast cancer. Breast Cancer Res. 2000, 2, 335–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersson, K.; Gustafsson, J.A. Role of estrogen receptor beta in estrogen action. Annu. Rev. Physiol. 2001, 63, 165–192. [Google Scholar] [CrossRef] [PubMed]
- Strauss, L.; Kallio, J.; Desai, N.; Pakarinen, P.; Miettinen, T.; Gylling, H.; Albrecht, M.; Makela, S.; Mayerhofer, A.; Poutanen, M. Increased exposure to estrogens disturbs maturation, steroidogenesis, and cholesterol homeostasis via estrogen receptor alpha in adult mouse Leydig cells. Endocrinology 2009, 150, 2865–2872. [Google Scholar] [CrossRef] [PubMed]
- Colasanti, T.; Maselli, A.; Conti, F.; Sanchez, M.; Alessandri, C.; Barbati, C.; Vacirca, D.; Tinari, A.; Chiarotti, F.; Giovannetti, A.; et al. Autoantibodies to estrogen receptor alpha interfere with T lymphocyte homeostasis and are associated with disease activity in systemic lupus erythematosus. Arthritis Rheum. 2012, 64, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Ribas, V.; Drew, B.G.; Le, J.A.; Soleymani, T.; Daraei, P.; Sitz, D.; Mohammad, L.; Henstridge, D.C.; Febbraio, M.A.; Hewitt, S.C.; et al. Myeloid-specific estrogen receptor alpha deficiency impairs metabolic homeostasis and accelerates atherosclerotic lesion development. Proc. Natl. Acad. Sci. USA 2011, 108, 16457–16462. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.C.; Cooney, A.J. The varied roles of nuclear receptors during vertebrate embryonic development. Nucl Recept Signal. 2003, 1. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Thompson, E.B. The structure of the nuclear hormone receptors. Steroids 1999, 64, 310–319. [Google Scholar] [CrossRef]
- Shiau, A.K.; Barstad, D.; Radek, J.T.; Meyers, M.J.; Nettles, K.W.; Katzenellenbogen, B.S.; Katzenellenbogen, J.A.; Agard, D.A.; Greene, G.L. Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism. Nat. Struct. Biol. 2002, 9, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Pike, A.C.; Brzozowski, A.M.; Hubbard, R.E.; Bonn, T.; Thorsell, A.G.; Engstrom, O.; Ljunggren, J.; Gustafsson, J.A.; Carlquist, M. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. EMBO J. 1999, 18, 4608–4618. [Google Scholar] [CrossRef] [PubMed]
- Paige, L.A.; Christensen, D.J.; Gron, H.; Norris, J.D.; Gottlin, E.B.; Padilla, K.M.; Chang, C.Y.; Ballas, L.M.; Hamilton, P.T.; McDonnell, D.P.; et al. Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta. Proc. Natl. Acad. Sci. USA 1999, 96, 3999–4004. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.K.; Smith, G.P. Searching for peptide ligands with an epitope library. Science 1990, 249, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Halachmi, S.; Marden, E.; Martin, G.; MacKay, H.; Abbondanza, C.; Brown, M. Estrogen receptor-associated proteins: Possible mediators of hormone-induced transcription. Science 1994, 264, 1455–1458. [Google Scholar] [CrossRef] [PubMed]
- Norris, J.D.; Paige, L.A.; Christensen, D.J.; Chang, C.Y.; Huacani, M.R.; Fan, D.; Hamilton, P.T.; Fowlkes, D.M.; McDonnell, D.P. Peptide antagonists of the human estrogen receptor. Science 1999, 285, 744–746. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, V.E.; Pedotti, M.; Chiado, A.; Simonelli, L.; Calzolai, L.; Varani, L.; Lettieri, T. Rational modification of estrogen receptor by combination of computational and experimental analysis. PLoS ONE 2014, 9, e102658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aqai, P.; Blesa, N.G.; Major, H.; Pedotti, M.; Varani, L.; Ferrero, V.E.; Haasnoot, W.; Nielen, M.W. Receptor-based high-throughput screening and identification of estrogens in dietary supplements using bioaffinity liquid-chromatography ion mobility mass spectrometry. Anal. Bioanal. Chem. 2013, 405, 9427–9436. [Google Scholar] [CrossRef] [PubMed]
- Usami, M.; Mitsunaga, K.; Ohno, Y. Estrogen receptor binding assay of chemicals with a surface plasmon resonance biosensor. J. Steroid Biochem. Mol. Biol. 2002, 81, 47–55. [Google Scholar] [CrossRef]
- Rich, R.L.; Hoth, L.R.; Geoghegan, K.F.; Brown, T.A.; LeMotte, P.K.; Simons, S.P.; Hensley, P.; Myszka, D.G. Kinetic analysis of estrogen receptor/ligand interactions. Proc. Natl. Acad. Sci. USA 2002, 99, 8562–8567. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, V.; Antonacci, A.; Patrolecco, L.; Lambreva, M.D.; Litescu, S.C.; Ghuge, S.A.; Rea, G. Analytical tools monitoring endocrine disrupting chemicals. TrAC Trends Anal. Chem. 2016, 80, 555–567. [Google Scholar] [CrossRef]
- Nguyen, H.; Park, J.; Kang, S.; Kim, M. Surface Plasmon Resonance: A Versatile Technique for Biosensor Applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-L.; Liu, Y.; Fan, T.; Hu, N.; Yang, Z.; Chen, X.; Wang, Z.-Y.; Yang, J. Design and Performance of a Portable and Multichannel SPR Device. Sensors 2017, 17, 1435. [Google Scholar] [CrossRef] [PubMed]
- Sumbayev, V.V.; Jensen, J.K.; Hansen, J.A.; Andreasen, P.A. Novel modes of oestrogen receptor agonism and antagonism by hydroxylated and chlorinated biphenyls, revealed by conformation-specific peptide recognition patterns. Mol. Cell. Endocrinol. 2008, 287, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Sumbayev, V.V.; Bonefeld-Jorgensen, E.C.; Wind, T.; Andreasen, P.A. A novel pesticide-induced conformational state of the oestrogen receptor ligand-binding domain, detected by conformation-specific peptide binding. FEBS Lett. 2005, 579, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, X.; Guo, L.H. Assessment of estrogenic activity of perfluoroalkyl acids based on ligand-induced conformation state of human estrogen receptor. Environ. Sci. Technol. 2013, 47, 634–641. [Google Scholar] [CrossRef] [PubMed]
- O’Shannessy, D.J.; Brigham-Burke, M.; Pek, K. Immobilization chemistries suitable for use in the BIAcore surface plasmon resonance detector. Anal. Biochem. 1992, 205, 132–136. [Google Scholar] [CrossRef]
- Pauwels, S.; Lintermans, A.; Neven, P.; Verhaeghe, J.; Jans, I.; Billen, J.; Vanderschueren, D.; Vermeersch, P. Need for estradiol assays with a lower functional sensitivity in clinical studies examining postmenopausal women treated with aromatase inhibitors. J. Clin. Oncol. 2013, 31, 509. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Bertin, J.; Gonthier, R.; Simard, J.-N.; Labrie, F. A sensitive, simple and robust LC–MS/MS method for the simultaneous quantification of seven androgen- and estrogen-related steroids in postmenopausal serum. J. Steroid Biochem. Mol. Biol. 2014, 144, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Hinteman, T.; Schneider, C.; Scholer, H.F.; Schneider, R.J. Field study using two immunoassays for the determination of estradiol and ethinylestradiol in the aquatic environment. Water Res. 2006, 40, 2287–2294. [Google Scholar] [CrossRef] [PubMed]
- Valdes, M.E.; Marino, D.J.; Wunderlin, D.A.; Somoza, G.M.; Ronco, A.E.; Carriquiriborde, P. Screening concentration of E1, E2 and EE2 in sewage effluents and surface waters of the “Pampas” region and the “Rio de la Plata” estuary (Argentina). Bull. Environ. Contam. Toxicol. 2015, 94, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Heub, S.; Tscharner, N.; Monnier, V.; Kehl, F.; Dittrich, P.S.; Follonier, S.; Barbe, L. Automated and portable solid phase extraction platform for immuno-detection of 17beta-estradiol in water. J. Chromatogr. A 2015, 1381, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Weis, K.E.; Ekena, K.; Thomas, J.A.; Lazennec, G.; Katzenellenbogen, B.S. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol. Endocrinol. 1996, 10, 1388–1398. [Google Scholar] [PubMed]
- Ozers, M.S.; Ervin, K.M.; Steffen, C.L.; Fronczak, J.A.; Lebakken, C.S.; Carnahan, K.A.; Lowery, R.G.; Burke, T.J. Analysis of ligand-dependent recruitment of coactivator peptides to estrogen receptor using fluorescence polarization. Mol. Endocrinol. 2005, 19, 25–34. [Google Scholar] [CrossRef] [PubMed]




© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
La Spina, R.; Ferrero, V.E.V.; Aiello, V.; Pedotti, M.; Varani, L.; Lettieri, T.; Calzolai, L.; Haasnoot, W.; Colpo, P. Label-Free Biosensor Detection of Endocrine Disrupting Compounds Using Engineered Estrogen Receptors. Biosensors 2018, 8, 1. https://0-doi-org.brum.beds.ac.uk/10.3390/bios8010001
La Spina R, Ferrero VEV, Aiello V, Pedotti M, Varani L, Lettieri T, Calzolai L, Haasnoot W, Colpo P. Label-Free Biosensor Detection of Endocrine Disrupting Compounds Using Engineered Estrogen Receptors. Biosensors. 2018; 8(1):1. https://0-doi-org.brum.beds.ac.uk/10.3390/bios8010001
Chicago/Turabian StyleLa Spina, Rita, Valentina E. V. Ferrero, Venera Aiello, Mattia Pedotti, Luca Varani, Teresa Lettieri, Luigi Calzolai, Willem Haasnoot, and Pascal Colpo. 2018. "Label-Free Biosensor Detection of Endocrine Disrupting Compounds Using Engineered Estrogen Receptors" Biosensors 8, no. 1: 1. https://0-doi-org.brum.beds.ac.uk/10.3390/bios8010001