Insects, Rodents, and Pets as Reservoirs, Vectors, and Sentinels of Antimicrobial Resistance
Abstract
:1. Introduction
2. Antimicrobial Resistance
2.1. Nature
2.2. Mechanisms of AMR
2.3. Conventional and Emerging Analytical Methods for Antimicrobial Resistance
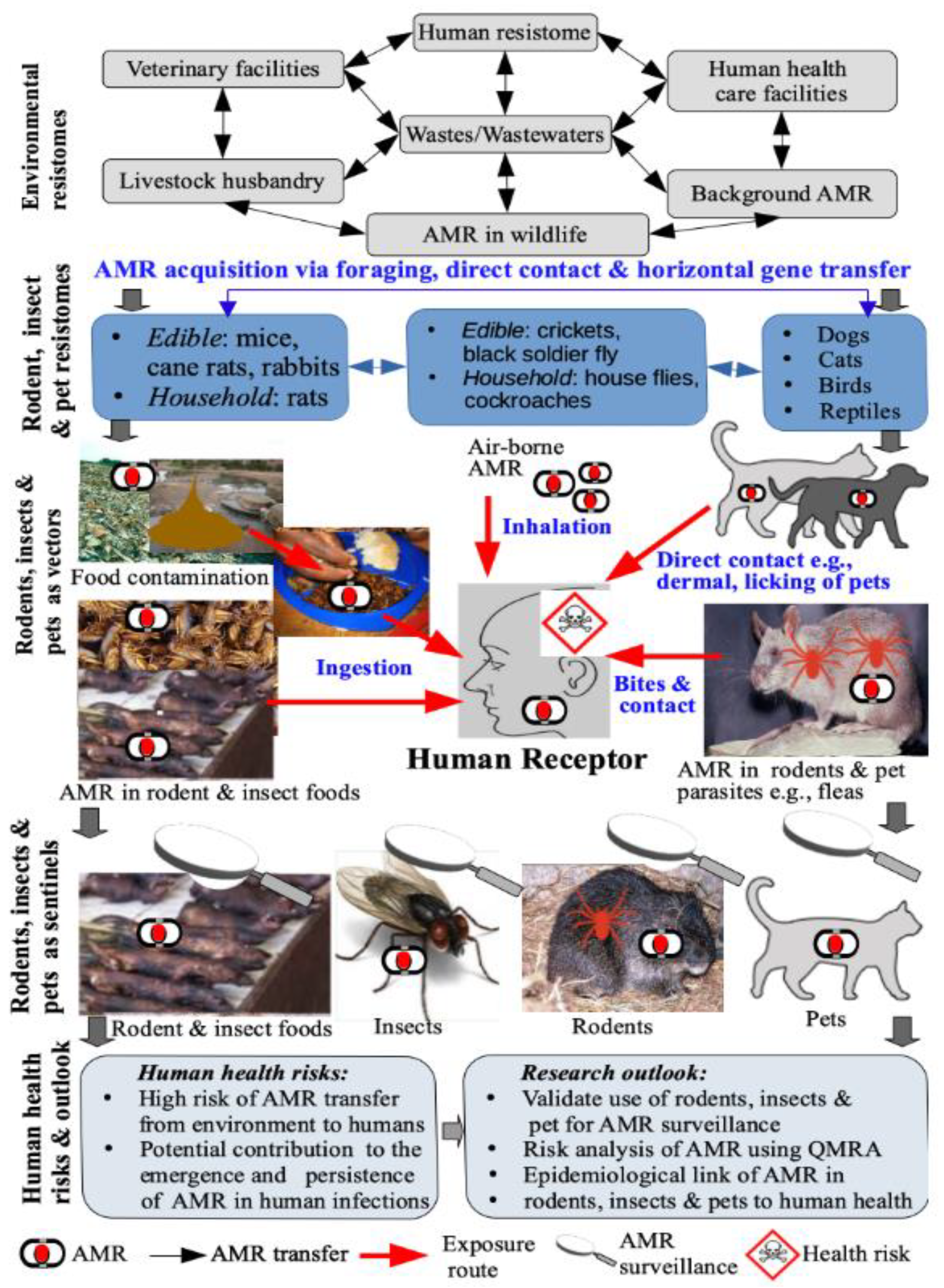
3. The Role of Insects, Rodents, and Pets in AMR Persistence and Transmission
3.1. Insects
3.1.1. How Insects Acquire Resistant Bacteria
3.1.2. Insects as Reservoirs of Antimicrobial Resistance
3.1.3. Insects as Vectors of AMR
3.1.4. Insects as Sentinels of AMR
3.2. Rodents
3.2.1. Rodents as Reservoirs of AMR
3.2.2. Rodents as Vectors of AMR Transmission
3.2.3. Rodents as Sentinels of AMR
3.3. Companion Animals/Pets
3.3.1. Pets as Reservoirs of AMR
3.3.2. Pets as Vectors and Sentinels of AMR
4. Human Exposure and Health Risks
4.1. Human Exposure Pathways
4.2. A Summary of the Inferential Evidence Pointing to Potential Human Health Risks
4.3. Drought on Fertile Grounds? AMR in Insects, Rodents, and Pets in Developing Countries
4.4. Towards a Quantitative Human Health Risk Assessment
5. Human Health Risk Assessment and Mitigation
5.1. Health Risk Assessment
- monitoring the usage of antimicrobial in companion animals,
- the extent to which the AMR occurs in pets, insects, and rodents entering households unintentionally,
- the association between AMR in rodents, insects, and animals of interest, and humans coming in contact with such animals, and
- the routes via which AMR microorganisms and/or ARGs can be transmitted between household animals and humans.
5.2. Mitigation
- limiting the use of antimicrobials in pets only to the clinically justified applications
- raising the awareness of the complex AMR issue within the veterinary specialists and even among the pet owners
- limiting direct contact between pet owners and animals during the use of antimicrobials
- using hand hygiene practices after direct contact with pets
- avoiding intimate contacts with pets through face licking and sharing a bed
- regular cleaning of households; and
- wearing rubber, latex, or vinyl gloves when cleaning urine and droppings from insects, rodents, and pets
- blocking all potential entry routes (foundation cracks, unsealed windows, doors, etc.), particularly in the colder season when the risk of entering is the highest
- sealing garbage bins and containers
- sealing food, including pet foods, to avoid cross-contamination
- cleaning the household to remove uneaten parts of food
- using traps and baits in case of a high risk of infestation; and
- preventing household cats from going outside to limit the predation, potential rodent-cat transmission, and further cat-human transmission.
- collection of dog’s and cat’s droppings and their appropriate disposal to avoid contact with insects (e.g., flies) that feed or develop in excrements, acquisition of AMR and its further spread to humans [95].
- regular cleaning of households
- The use of preventive measures to limit insects from entering the household setting (e.g., screens on windows, mosquito nets); and
- elimination of insects in household setting (e.g., mechanically or chemically in case of infestations)
6. Future Perspectives
6.1. Future Research Directions
6.1.1. Comprehensive Database on AMR in Insects, Rodents, and Companion Animals
6.1.2. Partitioning AMR between Natural and Anthropogenic Pools
6.1.3. Transfer Mechanisms and Behavior in the Environment–Animal–Human Interface
6.1.4. AMR Receptors and Primers in Insects, Rodents, and Companion Animals
6.1.5. Insects as Potential Sources of Novel Antimicrobials to Mitigate AMR
6.1.6. Human Exposure and Health Risks via the Consumption of Edible Insects and Rodents
6.1.7. Quantitative Microbial Risk Assessment
6.1.8. The Contribution of Insects, Rodents, and Pets to the Global Human AMR Burden
6.1.9. Understanding the ‘Human Factor’ in AMR
6.1.10. Increasing the Global Footprint of Developing Regions in AMR Research
6.2. Harnessing Emerging and Novel Tools to Unravel the Complex Behavior of AMR
6.2.1. Genomic Tools
6.2.2. Computational or In-Silico Techniques
6.2.3. Network Analysis
6.2.4. Big Data Analytics
7. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berry, D.B.; Lu, D.; Geva, M.; Watts, J.C.; Bhardwaj, S.; Oehler, A.; Renslo, A.R.; DeArmond, S.J.; Prusiner, S.B.; Giles, K. Drug resistance confounding prion therapeutics. Proc. Natl. Acad. Sci. USA 2013, 110, E4160–E4169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; The Review on Antimicrobial Resistance: London, UK, 2014; Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper (accessed on 28 November 2020).
- Dunachie, S.J.; Day, N.P.; Dolecek, C. The challenges of estimating the human global burden of disease of antimicrobial resistant bacteria. Curr. Opin. Microbiol. 2020, 57, 95–101. [Google Scholar] [CrossRef] [PubMed]
- GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Ma, Y.X.; Wang, C.Y.; Li, Y.Y.; Li, J.; Wan, Q.Q.; Chen, J.H.; Tay, F.R.; Niu, L.N. Considerations and caveats in combating ESKAPE pathogens against nosocomial infections. Adv. Sci. 2020, 7, 1901872. [Google Scholar] [CrossRef] [Green Version]
- Jiang, A.; Liu, N.; Said, R.A.; Ren, M.-D.; Gao, H.; Zheng, X.; Fu, X.; Liang, X.; Ruan, Z.-P.; Yao, Y.; et al. Nosocomial infections in gastrointestinal cancer patients: Bacterial profile, antibiotic resistance pattern, and prognostic factors. Cancer Manag. Res. 2020, 12, 4969–4979. [Google Scholar] [CrossRef]
- Ducey, T.; Durso, L.M.; Ibekwe, A.M.; Dungan, R.S.; Jackson, C.R.; Frye, J.G.; Castleberry, B.L.; Rashash, D.M.; Rothrock, M.J.; Boykin, D.; et al. A newly developed Escherichia coli isolate panel from a cross section of U.S. animal production systems reveals geographic and commodity-based differences in antibiotic resistance gene carriage. J. Hazard. Mater. 2020, 382, 120991. [Google Scholar] [CrossRef]
- Gwenzi, W.; Musiyiwa, K.; Mangori, L. Sources, behaviour and health risks of antimicrobial resistance genes in wastewaters: A hotspot reservoir. J. Environ. Chem. Eng. 2020, 8, 102220. [Google Scholar] [CrossRef]
- Laconi, A.; Mughini-Gras, L.; Tolosi, R.; Grilli, G.; Trocino, A.; Carraro, L.; Di Cesare, F.; Cagnardi, P.; Piccirillo, A. Microbial community composition and antimicrobial resistance in agricultural soils fertilized with livestock manure from conventional farming in Northern Italy. Sci. Total Environ. 2020, 143404. [Google Scholar] [CrossRef] [PubMed]
- Bueno, I.; Verdugo, C.; Jimenez-Lopez, O.; Alvarez, P.P.; Gonzalez-Rocha, G.; Lima, C.A.; Travis, D.A.; Wass, B.; Zhang, Q.; Ishii, S.; et al. Role of wastewater treatment plants on environmental abundance of Antimicrobial Resistance Genes in Chilean rivers. Int. J. Hyg. Environ. Health 2020, 223, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Gwenzi, W. The ‘thanato-resistome’—The funeral industry as a potential reservoir of antibiotic resistance: Early insights and perspectives. Sci. Total Environ. 2020, 749, 141120. [Google Scholar] [CrossRef]
- Sanganyado, E.; Gwenzi, W. Antibiotic resistance in drinking water systems: Occurrence, removal, and human health risks. Sci. Total Environ. 2019, 669, 785–797. [Google Scholar] [CrossRef]
- Xie, S.; Gu, A.Z.; Cen, T.; Li, D.; Chen, J. The effect and mechanism of urban fine particulate matter (PM2.5) on horizontal transfer of plasmid-mediated antimicrobial resistance genes. Sci. Total Environ. 2019, 683, 116–123. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, Z.; Hou, J.; Mao, D.; Lin, H.; Xue, Y.; Luo, Y. Monitoring antibiotic resistomes and bacterial microbiomes in the aerosols from fine, hazy, and dusty weather in Tianjin, China using a developed high-volume tandem liquid impinging sampler. Sci. Total Environ. 2020, 731, 139242. [Google Scholar] [CrossRef] [PubMed]
- Cenci-Goga, B.; Sechi, P.; Karama, M.; Ciavarella, R.; Pipistrelli, M.V.; Goretti, E.; Elia, A.C.; Gardi, T.; Pallottini, M.; Rossi, R.; et al. Cross-sectional study to identify risk factors associated with the occurrence of antimicrobial resistance genes in honey bees Apis mellifera) in Umbria, Central Italy. Environ. Sci. Pollut. Res. 2020, 27, 9637–9645. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.-S.; Li, Y.-Z.; Ge, J.; Xiao, G.; Mo, Y.; Wen, Y.-Q.; Liu, J.-P.; Xiong, Y.-Q.; Qiu, M.; Huo, S.-T.; et al. Comparisons of microbiological characteristics and antibiotic resistance of Klebsiella pneumoniae isolates from urban rodents, shrews, and healthy people. BMC Microbiol. 2020, 20, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obeng-Nkrumah, N.; Labi, A.-K.; Blankson, H.; Awuah-Mensah, G.; Oduro-Mensah, D.; Anum, J.; Teye, J.; Kwashie, S.D.; Bako, E.; Ayeh-Kumi, P.F.; et al. Household cockroaches carry CTX-M-15-, OXA-48- and NDM-1-producing enterobacteria, and share beta-lactam resistance determinants with humans. BMC Microbiol. 2019, 19, 272. [Google Scholar] [CrossRef]
- Odetoyin, B.W.; Adeola, B.; Olaniran, O. Frequency and antimicrobial resistance patterns of bacterial species isolated from the body surface of the housefly (Musca domestica) in Akure, Ondo State, Nigeria. J. Arthropod Borne Dis. 2020, 14, 88–96. [Google Scholar] [CrossRef] [Green Version]
- Desvars-Larrive, A.; Ruppitsch, W.; Lepuschitz, S.; Szostak, M.P.; Spergser, J.; Feßler, A.T.; Schwarz, S.; Monecke, S.; Ehricht, R.; Walzer, C.; et al. Urban brown rats (Rattus norvegicus) as possible source of multidrug-resistant Enterobacteriaceae and meticillin-resistant Staphylococcus spp., Vienna, Austria, 2016 and 2017. Eurosurveillance 2019, 24, 1900149. [Google Scholar] [CrossRef] [Green Version]
- Belas, A.; Marques, C.; Pomba, C. The gut microbiome and antimicrobial resistance in companion animals. In Advances in Animal Health, Medicine and Production; Duarte, A.F., Lopez de Costa, L., Eds.; Springer Science and Business Media: Berlin, Germany, 2020; pp. 233–245. [Google Scholar]
- Baldi, M.; Barquero-Calvo, E.; Hutter, S.E.; Walzer, C. Salmonellosis detection and evidence of antibiotic resistance in an urban raccoon population in a highly populated area, Costa Rica. Zoonoses Public Health 2019, 66, 852–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joosten, P.; Ceccarelli, D.; Odent, E.; Sarrazin, S.; Graveland, H.; van Gompel, L.; Battisti, A.; Caprioli, A.; Franco, A.; Wagenaar, J.A.; et al. Antimicrobial usage and resistance in companion animals: A cross-sectional study in three European countries. Antibiotics 2020, 9, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riwu, K.H.P.; Effendi, M.H.; Rantam, F.A. A review of extended spectrum β-lactamase (ESBL) producing Klebsiella pneumoniae and multidrug resistant (MDR) on companion animals. Syst. Rev. Pharm. 2020, 11, 270–277. [Google Scholar]
- Mathur, S.; Singh, R. Antibiotic resistance in food lactic acid bacteria—A review. Int. J. Food Microbiol. 2005, 105, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Morio, F.; Jensen, R.H.; Le Pape, P.; Arendrup, M.C. Molecular basis of antifungal drug resistance in yeasts. Int. J. Antimicrob. Agents 2017, 50, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Hokken, M.W.J.; Zwaan, B.; Melchers, W.; Verweij, P. Facilitators of adaptation and antifungal resistance mechanisms in clinically relevant fungi. Fungal Genet. Biol. 2019, 132, 103254. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 2016, 7, 1728. [Google Scholar] [CrossRef] [Green Version]
- Chou, S. Advances in the genotypic diagnosis of cytomegalovirus antiviral drug resistance. Antivir. Res. 2020, 176, 104711. [Google Scholar] [CrossRef]
- Guermouche, H.; Burrel, S.; Mercier-Darty, M.; Kofman, T.; Rogier, O.; Pawlotsky, J.-M.; Boutolleau, D.; Rodriguez, C. Characterization of the dynamics of human cytomegalovirus resistance to antiviral drugs by ultra-deep sequencing. Antivir. Res. 2020, 173, 104647. [Google Scholar] [CrossRef]
- Bergeron, S.; Raj, B.; Nathaniel, R.; Corbin, A.; LaFleur, G. Presence of antibiotic resistance genes in raw source water of a drinking water treatment plant in a rural community of USA. Int. Biodeterior. Biodegrad. 2017, 124, 3–9. [Google Scholar] [CrossRef]
- Hiller, C.; Hübner, U.; Fajnorova, S.; Schwartz, T.; Drewes, J.E. Antibiotic microbial resistance (AMR) removal efficiencies by conventional and advanced wastewater treatment processes: A review. Sci. Total Environ. 2019, 685, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, B.; Zhang, T. New insights into antibiotic resistome in drinking water and management perspectives: A metagenomic based study of small-sized microbes. Water Res. 2019, 152, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Hassell, J.M.; Ward, M.J.; Muloi, D.; Bettridge, J.M.; Robinson, T.P.; Kariuki, S.; Ogendo, A.; Kiiru, J.; Imboma, T.; Kang’ethe, E.K.; et al. Clinically relevant antimicrobial resistance at the wildlife–livestock–human interface in Nairobi: An epidemiological study. Lancet Planet. Health 2019, 3, E259–E269. [Google Scholar] [CrossRef] [Green Version]
- Furness, L.E.; Campbell, A.; Zhang, L.; Gaze, W.H.; McDonald, R.A. Wild small mammals as sentinels for the environmental transmission of antimicrobial resistance. Environ. Res. 2017, 154, 28–34. [Google Scholar] [CrossRef]
- Poudel, A.; Kang, Y.; Mandal, R.K.; Kalalah, A.; Butaye, P.; Hathcock, T.; Kelly, P.; Walz, P.; Macklin, K.; Cattley, R.; et al. Comparison of microbiota, antimicrobial resistance genes and mobile genetic elements in flies and the feces of sympatric animals. FEMS Microbiol. Ecol. 2020, 96, fiaa027. [Google Scholar] [CrossRef]
- Macovei, L.; Zurek, L. Ecology of antibiotic resistance genes: Characterization of enterococci from houseflies collected in food settings. Appl. Environ. Microbiol. 2006, 72, 4028–4035. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, A.; Ghosh, A.; Schal, C.; Zurek, L. Insects in confined swine operations carry a large antibiotic resistant and potentially virulent enterococcal community. BMC Microbiol. 2011, 11, 23. [Google Scholar] [CrossRef] [Green Version]
- Saidi, B.; Mafirakureva, P.; Mbanga, J. Antimicrobial resistance of Escherichia coli isolated from chickens with colibacillosis in and around Harare, Zimbabwe. Avian Dis. 2013, 57, 152–154. [Google Scholar] [CrossRef]
- Kmet, V.; Čuvalová, A.; Stanko, M. Small mammals as sentinels of antimicrobial-resistant staphylococci. Folia Microbiol. 2018, 63, 665–668. [Google Scholar] [CrossRef]
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef]
- Le, T.-H.; Ng, C.; Tran, N.H.; Chen, H.; Gin, K.Y.-H. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Res. 2018, 145, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Pai, H.-H.; Chen, W.-C.; Peng, C.-F. Isolation of bacteria with antibiotic resistance from household cockroaches (Periplaneta americana and Blattella germanica). Acta Trop. 2005, 93, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Pai, H.-H. Multidrug resistant bacteria isolated from cockroaches in long-term care facilities and nursing homes. Acta Trop. 2013, 125, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Nasirian, H. Infestation of cockroaches (Insecta: Blattaria) in the human dwelling environments: A systematic review and meta-analysis. Acta Trop. 2017, 167, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Oyeyemi, O.T.; Agbaje, M.O.; Okelue, U.B. Food-borne human parasitic pathogens associated with household cockroaches and houseflies in Nigeria. Parasite Epidemiol. Control 2016, 1, 10–13. [Google Scholar] [CrossRef] [Green Version]
- Guiyoule, A.; Gerbaud, G.; Buchrieser, C.; Galimand, M.; Rahalison, L.; Chanteau, S.; Courvalin, P.; Carnie, E. Transferrable plasmid-mediated resistance to streptomycin in a clinical isolate of Yersinia pestis. Emerg. Infect. Dis. 2001, 7, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Courvalin, P. Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 1994, 38, 1447–1451. [Google Scholar] [CrossRef] [Green Version]
- Perry, J.A.; Wright, G.D. The antibiotic resistance “mobilome”: Searching for the link between environment and clinic. Front. Microbiol. 2013, 4, 138. [Google Scholar] [CrossRef] [Green Version]
- Zurek, L.; Ghosh, A. Insects represent a link between food animal farms and the urban environment for antibiotic resistance traits. Appl. Environ. Microbiol. 2014, 80, 3562–3567. [Google Scholar] [CrossRef] [Green Version]
- Galimand, M.; Guiyoule, A.; Gerbaud, G.; Rasoamanana, B.; Chanteau, S.; Carniel, E.; Courvalin, P. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N. Engl. J. Med. 1997, 337, 677–681. [Google Scholar] [CrossRef]
- Doucet-Populaire, F.; Trieu-Cuot, P.; Dosbaa, I.; Andremont, A.; Courvalin, P. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob. Agents Chemother. 1991, 35, 185–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lester, C.H.; Frimodt-Møller, N.; Sørensen, T.L.; Monnet, D.L.; Hammerum, A.M. In vivo transfer of the vanA resistance gene from an Enterococcus faecium isolate of animal origin to an E. faecium isolate of human origin in the intestines of human volunteers. Antimicrob. Agents Chemother. 2006, 50, 596–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warnes, S.L.; Highmore, C.J.; Keevil, C.W. Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces: Implications for public health. mBio 2012, 3, 00489-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, P.R.; Gwanzura, L.; Latif, A.S.; Marowa, E.; Ray, S.; Katzenstein, D.A. Antimicrobial resistance in gonococci isolated from patients and from commercial sex workers in Harare, Zimbabwe. Int. J. Antimicrob. Agents 1998, 9, 175–179. [Google Scholar] [CrossRef]
- Guzman, J.; Vilcinskas, A. Bacteria associated with cockroaches: Health risk or biotechnological opportunity? Appl. Microbiol. Biotechnol. 2020, 104, 10369–10387. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Cloud-Hansen, K.A.; Wolinski, J.M.; Guan, C.; Greene, S.; Lu, S.; Boeyink, M.; Broderick, N.A.; Raffa, K.F.; Handelsman, J. Resident microbiota of the gypsy moth midgut harbors antibiotic resistance determinants. DNA Cell Biol. 2009, 28, 109–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolejska, M.; Literak, I. Wildlife Is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 2019, 63, 01167-19. [Google Scholar] [CrossRef] [Green Version]
- Ahlstrom, C.A.; Bonnedahl, J.; Woksepp, H.; Hernandez, J.; Olsen, B.; Ramey, A.M. Acquisition and dissemination of cephalosporin-resistant E. coli in migratory birds sampled at an Alaska landfill as inferred through genomic analysis. Sci. Rep. 2018, 8, 7361. [Google Scholar] [CrossRef]
- Skarżyńska, M.; Zając, M.; Kamińska, E.; Bomba, A.; Żmudzki, J.; Jabłoński, A.; Wasyl, D. Salmonella and antimicrobial resistance in wild rodents—True or false threat? Pathogens 2020, 9, 771. [Google Scholar] [CrossRef]
- Tunstall, T.; Portelli, S.; Phelan, J.; Clark, T.G.; Ascher, D.B.; Furnham, N. Combining structure and genomics to understand antimicrobial resistance. Comput. Struct. Biotechnol. J. 2020, 18, 3377–3394. [Google Scholar] [CrossRef]
- Lv, J.; Deng, S.; Zhang, L. A review of artificial intelligence applications for antimicrobial resistance. Biosaf. Health 2020. [Google Scholar] [CrossRef]
- Garcia-Vidal, C.; Sanjuan, G.; Puerta-Alcalde, P.; Moreno-García, E.; Soriano, A. Artificial intelligence to support clinical decision-making processes. EBioMedicine 2019, 46, 27–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.P.; Wang, H.; Durant, T.J.; Mathison, B.A.; Sharp, S.E.; Kirby, J.E.; Long, S.W.; Rhoads, D.D. Applications of artificial intelligence in clinical microbiology diagnostic testing. Clin. Microbiol. Newsl. 2020, 42, 61–70. [Google Scholar] [CrossRef]
- Pava-Ripoll, M.; Pearson, R.E.G.; Miller, A.K.; Ziobro, G.C. Prevalence and relative risk of Cronobacter spp., Salmonella spp., and Listeria monocytogenes associated with the body surfaces and guts of individual filth flies. Appl. Environ. Microbiol. 2012, 78, 7891–7902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usui, M.; Iwasa, T.; Fukuda, A.; Sato, T.; Okubo, T.; Tamura, Y. The role of flies in spreading the extended-spectrum β-lactamase gene from cattle. Microb. Drug Resist. 2013, 19, 415–420. [Google Scholar] [CrossRef]
- Graham, J.; Price, L.B.; Evans, S.L.; Graczyk, T.K.; Silbergeld, E.K. Antibiotic resistant enterococci and staphylococci isolated from flies collected near confined poultry feeding operations. Sci. Total Environ. 2009, 407, 2701–2710. [Google Scholar] [CrossRef]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef]
- Collignon, P.J.; McEwen, S.A. One health—Its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, A.; Usui, M.; Okubo, T.; Tagaki, C.; Sukpanyatham, N.; Tamura, Y. Co-harboring of cephalosporin (bla)/colistin (mcr) resistance genes among Enterobacteriaceae from flies in Thailand. FEMS Microbiol. Lett. 2018, 365, 1–7. [Google Scholar] [CrossRef]
- Sobur, A.; Haque, Z.F.; Sabuj, A.A.; Ievy, S.; Rahman, A.T.; El Zowalaty, M.E.; Rahman, T. Molecular detection of multidrug and colistin-resistant Escherichia coli isolated from house flies in various environmental settings. Futur. Microbiol. 2019, 14, 847–858. [Google Scholar] [CrossRef]
- Tufa, T.B.; Fuchs, A.; Wienemann, T.; Eggers, Y.; Abdissa, S.; Schneider, M.; Jensen, B.-E.O.; Bode, J.G.; Pfeffer, K.; Häussinger, D.; et al. Carriage of ESBL-producing Gram-negative bacteria by flies captured in a hospital and its suburban surroundings in Ethiopia. Antimicrob. Resist. Infect. Control. 2020, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Graczyk, T.K.; Knight, R.; Gilman, R.H.; Cranfield, M.R. The role of non-biting flies in the epidemiology of human infectious diseases. Microbes Infect. 2001, 3, 231–235. [Google Scholar] [CrossRef]
- Graczyk, T.K.; Cranfield, M.R.; Bixler, H.; Fayer, R. House flies (Musca domestica) as transport hosts of Cryptosporidium parvum. Am. J. Trop. Med. Hyg. 1999, 61, 500–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, J. Adaptive features on the tarsi of cockroaches (Insecta: Dictyoptera). Int. J. Insect Morphol. Embryol. 1974, 3, 317–334. [Google Scholar] [CrossRef]
- Yap, K.L.; Kalpana, M.; Lee, H.L. Wings of the common house fly (Musca domestica L.): Importance in mechanical transmission of Vibrio cholerae. Trop. Biomed. 2008, 25, 1–8. [Google Scholar]
- Fathpour, H.; Emtiazi, G.; Ghasemi, E. Cockroaches as reservoirs and vectors of drug resistant Salmonella spp. Fresenius Environ. Bull. 2003, 12, 724–727. [Google Scholar]
- Wang, H.; Sangwan, N.; Li, H.-Y.; Su, J.-Q.; Oyang, W.-Y.; Zhang, Z.-J.; Gilbert, J.A.; Zhu, Y.-G.; Ping, F.; Zhang, H.-L. The antibiotic resistome of swine manure is significantly altered by association with the Musca domestica larvae gut microbiome. ISME J. 2017, 11, 100–111. [Google Scholar] [CrossRef]
- Wang, H.; Li, H.; Gilbert, J.A.; Li, H.; Wu, L.; Liu, M.; Wang, L.; Zhou, Q.; Yuan, J.; Zhang, Z. Housefly larva vermicomposting efficiently attenuates antibiotic resistance genes in swine manure, with concomitant bacterial population changes. Appl. Environ. Microbiol. 2015, 81, 7668–7679. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, A.; Usui, M.; Okamura, M.; Dong-Liang, H.; Tamura, Y. Role of flies in the maintenance of antimicrobial resistance in farm environments. Microb. Drug Resist. 2019, 25, 127–132. [Google Scholar] [CrossRef]
- Nazni, W.A.; Luke, H.; Rozita, W.M.W.; Abdullah, A.G.; Sa’Diyah, I.; Azahari, A.H.; Zamree, I.; Tan, S.B.; Lee, H.L.; Sofian, M.A. Determination of the flight range and dispersal of the house fly, Musca domestica (L.) using mark release recapture technique. Trop. Biomed. 2005, 22, 53–61. [Google Scholar]
- Tian, B.; Fadhil, N.H.; Powell, J.E.; Kwong, W.K.; Moran, N.A. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 2012, 3, 4–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ignasiak, K.; Maxwell, A. Antibiotic-resistant bacteria in the guts of insects feeding on plants: Prospects for discovering plant-derived antibiotics. BMC Microbiol. 2017, 17, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubert, J.; Stejskal, V.; Athanassiou, C.; Throne, J.E. Health hazards associated with arthropod infestation of stored products. Annu. Rev. Entomol. 2018, 63, 553–573. [Google Scholar] [CrossRef] [PubMed]
- Doggett, S.L.; Dwyer, D.E.; Peñas, P.F.; Russell, R.C. Bed bugs: Clinical relevance and control options. Clin. Microbiol. Rev. 2012, 25, 164–192. [Google Scholar] [CrossRef] [Green Version]
- Boiocchi, F.; Davies, M.P.; Hilton, A.C. An examination of flying insects in seven hospitals in the United Kingdom and carriage of bacteria by true flies (Diptera: Calliphoridae, Dolichopodidae, Fanniidae, Muscidae, Phoridae, Psychodidae, Sphaeroceridae). J. Med Entomol. 2019, 56, 1684–1697. [Google Scholar] [CrossRef]
- Anacarso, I.; Iseppi, R.; Sabia, C.; Messi, P.; Condò, C.; Bondi, M.; De Niederhäusern, S. Conjugation-mediated transfer of antibiotic-resistance plasmids between Enterobacteriaceae in the digestive tract of Blaberus craniifer (Blattodea: Blaberidae). J. Med Entomol. 2016, 53, 591–597. [Google Scholar] [CrossRef]
- Fukuda, A.; Usui, M.; Okubo, T.; Tamura, Y. Horizontal transfer of plasmid-mediated cephalosporin resistance genes in the intestine of houseflies (Musca domestica). Microb. Drug Resist. 2016, 22, 336–341. [Google Scholar] [CrossRef]
- Akhtar, M.; Hirt, H.; Zurek, L. Horizontal transfer of the tetracycline resistance gene tetM mediated by pCF10 among Enterococcus faecalis in the house fly (Musca domestica L.) alimentary canal. Microb. Ecol. 2009, 58, 509–518. [Google Scholar] [CrossRef]
- Petridis, M.; Bagdasarian, M.; Waldor, M.K.; Walker, E. Horizontal transfer of Shiga toxin and antibiotic resistance genes among Escherichia coli strains in house fly (Diptera: Muscidae) gut. J. Med. Entomol. 2006, 43, 288–295. [Google Scholar] [CrossRef]
- Cotton, M.; Wasserman, E.; Pieper, C.; Theron, D.; Tubbergh, D.; Campbell, G.; Fang, F.; Barnes, A.J. Invasive disease due to extended spectrum beta-lactamase-producing Klebsiella pneumoniae in a neonatal unit: The possible role of cockroaches. J. Hosp. Infect. 2000, 44, 13–17. [Google Scholar] [CrossRef]
- Farag, T.H.; Faruque, A.S.; Wu, Y.; Das, S.K.; Hossain, A.; Ahmed, S.; Ahmed, D.; Nasrin, D.; Kotloff, K.L.; Panchilangam, S.; et al. Housefly population density correlates with shigellosis among children in Mirzapur, Bangladesh: A time series analysis. PLoS Negl. Trop. Dis. 2013, 7, e2280. [Google Scholar] [CrossRef] [PubMed]
- De Jesús, A.J.; Olsen, A.R.; Bryce, J.R.; Whiting, R.C. Quantitative contamination and transfer of Escherichia coli from foods by houseflies, Musca domestica L. (Diptera: Muscidae). Int. J. Food Microbiol. 2004, 93, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Onwugamba, F.C.; Fitzgerald, J.R.; Rochon, K.; Guardabassi, L.; Alabi, A.; Kühne, S.; Grobusch, M.P.; Schaumburg, F. The role of ‘filth flies’ in the spread of antimicrobial resistance. Travel Med. Infect. Dis. 2018, 22, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Stoffolano, J.G. Fly foregut and transmission of microbes. Adv. Insect Physiol. 2019, 57, 27–95. [Google Scholar] [CrossRef]
- Doud, C.W.; Zurek, L. Enterococcus faecalis OG1RF:pMV158 survives and proliferates in the house fly digestive tract. J. Med Entomol. 2012, 49, 150–155. [Google Scholar] [CrossRef] [Green Version]
- Allen, B. Excretion of viable tubercle bacilli by Blatta orientalis (the oriental cockroach) following ingestion of heat-fixed sputum smears: A laboratory investigation. Trans. R. Soc. Trop. Med. Hyg. 1987, 81, 98–99. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Maritz, J.M.; Carlton, J.M.; Schal, C. Overlapping community compositions of gut and fecal microbiomes in lab-reared and field-collected german cockroaches. Appl. Environ. Microbiol. 2018, 84, e01037-18. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.-K.; Yong, H.I.; Kim, Y.-B.; Kim, H.-W.; Choi, Y.-S. Edible insects as a protein source: A review of public perception, processing technology, and research trends. Food Sci. Anim. Resour. 2019, 39, 521–540. [Google Scholar] [CrossRef] [Green Version]
- Levine, O.S.; Levine, M.M. Houseflies (Musca domestica) as mechanical vectors of shigellosis. Clin. Infect. Dis. 1991, 13, 688–696. [Google Scholar] [CrossRef]
- Kopanic, R.J.; Sheldon, B.W.; Wright, C.G. Cockroaches as vectors of salmonella: Laboratory and field trials. J. Food Prot. 1994, 57, 125–135. [Google Scholar] [CrossRef]
- Dunbar, J.P.; Khan, N.A.; Abberton, C.L.; Brosnan, P.; Murphy, J.; Afoullouss, S.; O’Flaherty, V.; Dugon, M.M.; Boyd, A. Synanthropic spiders, including the global invasive noble false widow Steatoda nobilis, are reservoirs for medically important and antibiotic resistant bacteria. Sci. Rep. 2020, 10, 20916. [Google Scholar] [CrossRef] [PubMed]
- Hyde, J.; Gorham, C.; Brackney, D.E.; Steven, B. Antibiotic resistant bacteria and commensal fungi are common and conserved in the mosquito microbiome. PLoS ONE 2019, 14, e0218907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, C.F.; Romney, M.G. Bedbugs as vectors for drug-resistant bacteria. Emerg. Infect. Dis. 2011, 17, 1132–1134. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.P.; Leibler, J.H.; Price, L.B.; Otte, J.M.; Pfeiffer, D.U.; Tiensin, T.; Silbergeld, E.K. The animal-human interface and infectious disease in industrial food animal production: Rethinking biosecurity and biocontainment. Public Health Rep. 2008, 123, 282–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matilla, F.; Velleman, Y.; Harrison, W.; Nevel, M. Animal influence on water, sanitation and hygiene measures for zoonosis control at the household level: A systematic literature review. PLoS Negl. Trop. Dis. 2018, 12, e0006619. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Yang, Y.; Wu, N.; Gao, M.; Tan, Y. Combined toxicity of pyrethroid insecticides and heavy metals: A review. Environ. Chem. Lett. 2019, 17, 1693–1706. [Google Scholar] [CrossRef]
- Yu, Y.; Li, X.; Yang, G.; Wang, Y.; Wang, X.; Cai, L.; Liu, X. Joint toxic effects of cadmium and four pesticides on the earthworm (Eisenia fetida). Chemosphere 2019, 227, 489–495. [Google Scholar] [CrossRef]
- Pu, Q.; Fan, X.-T.; Li, H.; An, X.-L.; Lassen, S.B.; Su, J.-Q. Cadmium enhances conjugative plasmid transfer to a fresh water microbial community. Environ. Pollut. 2021, 268, 115903. [Google Scholar] [CrossRef]
- Chelliah, R.; Park, B.-J.; Wei, S.; Park, J.-H.; Park, Y.-S.; Kim, S.-H.; Jin, Y.; Oh, D.-H. New perspectives on Mega plasmid sequence (poh1) in Bacillus thuringiensis ATCC 10792 harbouring antimicrobial, insecticidal and antibiotic resistance genes. Microb. Pathog. 2019, 126, 14–18. [Google Scholar] [CrossRef]
- Galimand, M.; Carniel, E.; Courvalin, P. Resistance of Yersinia pestis to antimicrobial agents. Antimicrob. Agents Chemother. 2006, 50, 3233–3236. [Google Scholar] [CrossRef] [Green Version]
- Hinnebusch, B.J.; Rosso, M.-L.; Schwan, T.G.; Carniel, E. High-frequency conjugative transfer of antibiotic resistance genes to Yersinia pestis in the flea midgut. Mol. Microbiol. 2002, 46, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Graczyk, T.K.; Knight, R.; Tamang, L. Mechanical transmission of human protozoan parasites by insects. Clin. Microbiol. Rev. 2005, 18, 128–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, A.A.; Skovgard, H.; Stockmarr, A.; Handberg, K.J.; Jorgensen, P.H. Persistence of low-pathogenic avian influenza H5N7 and H7N1 subtypes in house flies (Diptera: Muscidae). J. Med. Entomol. 2011, 48, 608–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davari, B.; Khodavaisy, S.; Ala, F. Isolation of fungi from housefly (Musca domestica L.) at Slaughter House and Hospital in Sanandaj, Iran. J. Prev. Med. Hyg. 2012, 53, 172–174. [Google Scholar]
- Kassiri, H.; Zarrin, M.; Veys-Behbahani, R.; Faramarzi, S.; Kasiri, A. Isolation and identification of pathogenic filamentous fungi and yeasts from adult house fly (Diptera: Muscidae) captured from the hospital environments in Ahvaz City, southwestern Iran. J. Med. Entomol. 2015, 52, 1351–1356. [Google Scholar] [CrossRef]
- Haddow, A.D.; Nasar, F.; Schellhase, C.W.; Moon, R.D.; Padilla, S.L.; Zeng, X.; Wollen, S.E.; Shamblin, J.D.; Grimes, E.C.; Zelko, J.M.; et al. Low potential for mechanical transmission of Ebola virus via house flies (Musca domestica). Parasites Vectors 2017, 10, 218. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ma, Z.-B.; Zeng, Z.-L.; Yang, X.-W.; Huang, Y.; Liu, J.-H. Response to comment on “The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes”. Zool. Res. 2017, 38, 212. [Google Scholar] [CrossRef] [Green Version]
- Ranjbar, R.; Izadi, M.; Hafshejani, T.T.; Khamesipour, F. Molecular detection and antimicrobial resistance of Klebsiella pneumoniae from house flies (Musca domestica) in kitchens, farms, hospitals and slaughterhouses. J. Infect. Public Health 2016, 9, 499–505. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-C.; Chang, Y.-C.; Chuang, H.-L.; Chiu, C.-C.; Lee, O.K.-S.; Chang, C.-C.; Hsuan, S.-L.; Lin, W.-H.; Chen, T.-H. Transmission of Salmonella between swine farms by the housefly (Musca domestica). J. Food Prot. 2011, 74, 1012–1016. [Google Scholar] [CrossRef]
- Olsen, A.R. Regulatory action criteria for filth and other extraneous materials. Regul. Toxicol. Pharmacol. 1998, 28, 199–211. [Google Scholar] [CrossRef] [Green Version]
- Stafford, K.C. Fly Management Handbook: A Guide to Biology, Dispersal, and Management of the House Fly and Related Flies for Farmers, Municipalities, and Public Health Officials; The Connecticut Agricultural Experiment Station: New Haven, CT, USA, 2008. [Google Scholar]
- Fotedar, R. Vector potential of houseflies (Musca domestica) in the transmission of Vibrio cholerae in India. Acta Trop. 2001, 78, 31–34. [Google Scholar] [PubMed]
- Agbodaze, D.; Owusu, S.B. Cockroaches (Periplaneta americana) as carriers of agents of bacterial diarrhoea in Accra, Ghana. Central Afr. J. Med. 1989, 35, 484–486. [Google Scholar]
- Fotedar, R.; Shriniwas, U.B.; Verma, A. Cockroaches (Blattella germanica) as carriers of microorganisms of medical importance in hospitals. Epidemiology Infect. 1991, 107, 181–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pai, H.; Chen, W.; Peng, C. Isolation of non-tuberculous mycobacteria from hospital cockroaches (Periplaneta americana). J. Hosp. Infect. 2003, 53, 224–228. [Google Scholar] [CrossRef]
- Rivault, C. Spatial distribution of the cockroach, Blattella germanica, in a swimming-bath facility. Entomol. Exp. Et Appl. 1989, 53, 247–255. [Google Scholar] [CrossRef]
- Rivault, C.; Cloarec, A.; Le Guyader, A. Bacterial contamination of food by cockroaches. J. Environ. Health 1993, 55, 21–23. [Google Scholar]
- Islam, S.; Nath, A.D.; Islam, K.; Chakma, S.; Hossain, M.B.; Al Faruq, A.; Hassan, M.M. Isolation, identification and antimicrobial resistance profile of Staphylococcus aureus in Cockroaches (Periplaneta americana). J. Adv. Vet. Anim. Res. 2016, 3, 221–228. [Google Scholar] [CrossRef]
- Menasria, T.; Moussa, F.; El-Hamza, S.; Tine, S.; Megri, R.; Chenchouni, H. Bacterial load of German cockroach (Blattella germanica) found in hospital environment. Pathog. Glob. Health 2014, 108, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Akinjogunla, O.J.; Odeyemi, A.; Udoinyang, E. Cockroaches (Periplaneta americana and Blattella germanica): Reservoirs of multi drug resistant (MDR) bacteria in Uyo, Akwa Ibom State. Sci. J. Biol. Sci. 2012, 1, 19–30. [Google Scholar]
- Vandeweyer, D.; Milanovic, V.; Garofalo, C.; Osimani, A.; Clementi, F.; Van Campenhout, L.; Acquilanti, L. Real-time PCR detection and quantification of selected transferable antibiotic resistance genes in fresh edible insects from Belgium and the Netherlands. Int. J. Food Microbiol. 2019, 290, 288–295. [Google Scholar] [CrossRef]
- Osimani, A.; Garofalo, C.; Aquilanti, L.; Milanovic, V.; Cardinali, F.; Taccari, M.; Pasquini, M.; Tavoletti, S. Transferable antibiotic resistances in marketed edible grasshoppers (Locusta migratoria migratorioides). J. Food Sci. 2017, 82, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Milanović, V.; Osimani, A.; Pasquini, M.; Aquilanti, L.; Garofalo, C.; Taccari, M.; Cardinali, F.; Riolo, P.; Clementi, F. Getting insight into the prevalence of antibiotic resistance genes in specimens of marketed edible insects. Int. J. Food Microbiol. 2016, 227, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Eutick, M.L.; O’Brien, R.W.; Slaytor, M. Bacteria from the gut of Australian termites. Appl. Environ. Microbiol. 1978, 35, 823–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlüter, O.; Rumpold, B.; Holzhauser, T.; Roth, A.; Vogel, R.F.; Quasigroch, W.; Vogel, S.; Heinz, V.; Jäger, H.; Bandick, N.; et al. Safety aspects of the production of foods and food ingredients from insects. Mol. Nutr. Food Res. 2017, 61, 1–14. [Google Scholar] [CrossRef]
- Van Huis, A. Edible insects are the future? Proc. Nutr. Soc. 2016, 75, 294–305. [Google Scholar] [CrossRef] [Green Version]
- Garofalo, C.; Osimani, A.; Milanovic, V.; Taccari, M.; Cardinali, F.; Aquilanti, L.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Clementi, F. The microbiota of marketed processed edible insects as revealed by high-throughput sequencing. Food Microbiol. 2017, 62, 15–22. [Google Scholar] [CrossRef]
- Vandeweyer, D.; Crauwels, S.; Lievens, B.; Van Campenhout, L. Metagenetic analysis of the bacterial communities of edible insects from diverse production cycles at industrial rearing companies. Int. J. Food Microbiol. 2017, 261, 11–18. [Google Scholar] [CrossRef]
- Klunder, H.; Wolkers-Rooijackers, J.; Korpela, J.; Nout, M. Microbiological aspects of processing and storage of edible insects. Food Control 2012, 26, 628–631. [Google Scholar] [CrossRef]
- Banjo, A.D.; Lawal, O.A.; Adeyemi, A.J. The microbial fauna associated with the larvae of Oryctes monocerus. J. Appl. Sci. Res. 2006, 2, 837–843. [Google Scholar]
- GeneWatch UK Report 2015. Genetically Modified Insect Factories: A New Source of Superbugs? Available online: http://tinyurl.com/omokzmk (accessed on 10 December 2020).
- Literák, I.; Dolejska, M.; Rybarikova, J.; Cizek, A.; Strejckova, P.; Vyskočilová, M.; Friedman, M.; Klimes, J. Highly variable patterns of antimicrobial resistance in commensal Escherichia coli Isolates from pigs, sympatric rodents, and flies. Microb. Drug Resist. 2009, 15, 229–237. [Google Scholar] [CrossRef]
- Channaiah, L.H.; Subramanyam, B.; McKinney, L.J.; Zurek, L. Stored-product insects carry antibiotic-resistant and potentially virulent enterococci. FEMS Microbiol. Ecol. 2010, 74, 464–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Channaiah, L.H.; Subramanyam, B.; Zurek, L. Survival of Enterococcus faecalis OG1RF:pCF10 in poultry and cattle feed: Vector competence of the red flour beetle, Tribolium castaneum (Herbst). J. Food Prot. 2010, 73, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Rodovalho, C.D.; Santos, A.L.; Marcolino, M.T.; Bonetti, A.M.; Brandeburgo, M.A.M. Urban ants and transportation of nosocomial bacteria. Neotrop. Entomol. 2007, 36, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Huff, R.; Pereira, R.I.; Pissetti, C.; de Araújo, A.M.; d’Azevedo, P.A.; Frazzon, J.; GuedesFrazzon, A.P. Antimicrobial resistance and genetic relationships of enterococci from siblings and non-siblings Heliconius erato phyllis caterpillars. PeerJ 2020, 8, e8647. [Google Scholar] [CrossRef] [PubMed]
- Pietri, J.E. Case not Closed: Arguments for New Studies of the Interactions between Bed Bugs and Human Pathogens. Am. J. Trop. Med. Hyg. 2020, 103, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Kruse, H.; Sørum, H. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 1994, 60, 4015–4021. [Google Scholar] [CrossRef] [Green Version]
- Butaye, P.; Argudín, M.A.; Threlfall, J. Introduction to Antimicrobial-Resistant Foodborne Pathogens; Elsevier BV: Amsterdam, The Netherlands, 2015; pp. 1–17. [Google Scholar]
- Huijbers, P.M.; Flach, C.-F.; Larsson, D.G.J. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 2019, 130, 104880. [Google Scholar] [CrossRef]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential impact of antimicrobial resistance in wildlife, environment and human health. Front. Microbiol. 2014, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Jardine, C.M.; Boerlin, P.; Janecko, N.; Lumsden, J.S.; Barker, I.K.; Pearl, D.L.; Reid-Smith, R.J.; Jardine, C. Antimicrobial resistance in generic Escherichia coli isolates from wild small mammals living in swine farm, residential, landfill, and natural environments in southern Ontario, Canada. Appl. Environ. Microbiol. 2010, 77, 882–888. [Google Scholar] [CrossRef] [Green Version]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 2008, 75, 559–566. [Google Scholar] [CrossRef] [Green Version]
- Himsworth, C.G.; Zabek, E.; Desruisseau, A.; Parmley, E.J.; Reid-Smith, R.; Leslie, M.; Ambrose, N.; Patrick, D.M.; Cox, W. Avian pathogenicity genes and antibiotic resistance in Escherichia coli isolates from wild Norway rats (Rattus norvegicus) in British Columbia, Canada. J. Wildl. Dis. 2016, 52, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Himsworth, C.G.; Miller, R.R.; Montoya, V.; Hoang, L.; Romney, M.G.; Al-Rawahi, G.N.; Kerr, T.; Jardine, C.M.; Patrick, D.M.; Tang, P.; et al. Carriage of methicillin-resistant Staphylococcus aureus by wild urban Norway rats (Rattus norvegicus). PLoS ONE 2014, 9, e87983. [Google Scholar] [CrossRef] [PubMed]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009, 35, 221–270. [Google Scholar] [CrossRef] [PubMed]
- Gilliver, M.A.; Bennetti, M.; Begon, M.; Hazel, S.M.; Hart, C.A. Antibiotic resistance in wild rodents. Nature 1999, 401, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Kayihura, M. Salmonella Reservoirs in Animals as Source of Infection. Master’s Thesis, Nairobi University, Nairobi, Kenya, 1982. [Google Scholar]
- Ong, K.H.; Khor, W.C.; Quek, J.Y.; Low, Z.X.; Arivalan, S.; Humaidi, M.; Chua, C.; Seow, K.L.G.; Guo, S.; Tay, M.Y.F.; et al. Occurrence and antimicrobial resistance traits of Escherichia coli from wild birds and rodents in Singapore. Int. J. Environ. Res. Public Health 2020, 17, 5606. [Google Scholar] [CrossRef] [PubMed]
- Cleri, D.J.; Varnaleo, J.R.; Lombardi, I.J.; Rabbat, M.S.; Matthew, A.; Marton, R.; Reyelt, M.C. Plague pneumonia caused by Yersinia Pestis. Semin. Respir. Infect. 1997, 12, 12–23. [Google Scholar]
- Wullenweber, M. Streptobacillus moniliformis—A zoonotic pathogen. Taxonomic considerations, host species, diagnosis, therapy and geographical distribution. J. Lab. Anim. Dis. 1995, 29, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Burt, S.A.; Siemeling, L.; Kuijper, E.J.; Lipman, L.J.A. Vermin on pig farms are vectors for Clostridium difficile PCR ribotypes 078 and 045. Vet. Microbiol. 2012, 160, 256–258. [Google Scholar] [CrossRef]
- Meerburg, B.G.; Jacobs-Reitsma, W.F.; Wagenaar, J.A.; Kijlstra, A. Presence of Salmonella and Campylobacter spp. in wild small mammals on bacteria and resistance genes in NYC house mice. Appl. Environ Microbiol. 2006, 72, 960–962. [Google Scholar] [CrossRef] [Green Version]
- Shimi, A.; Keyhani, M.; Hedayati, K. Studies on salmonellosis in the house mouse, Mus musculus. Lab. Anim. 1979, 13, 33–34. [Google Scholar] [CrossRef]
- Williams, S.H.; Che, X.; Paulick, A.; Guo, C.; Lee, B.; Muller, D.; Uhlemann, A.-C.; Lowy, F.D.; Corrigan, R.M.; Lipkin, W.I. New York City house mice (Mus musculus) as potential reservoirs for pathogenic bacteria and antimicrobial resistance determinants. mBio 2018, 9, e00624-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikesic, D.G.; Drickamer, L.C. Factors affecting home-range size in house mice (Mus musculus domesticus) living in outdoor enclosures. Am. Midl. Nat. 1992, 127, 31. [Google Scholar] [CrossRef]
- Courvalin, P. The Garrod Lecture Evasion of antibiotic action by bacteria. J. Antimicrob. Chemother. 1996, 37, 855–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, I. Penetration of antimicrobial to their target sites. J. Antimicrob. Chemother. 1990, 26, 607–608. [Google Scholar] [CrossRef] [Green Version]
- Ribas, A.; Saijuntha, W.; Agatsuma, T.; Prantlová, V.; Poonlaphdecha, S. Rodents as a source of Salmonella contamination in wet markets in Thailand. Vector Borne Zoonotic Dis. 2016, 16, 537–540. [Google Scholar] [CrossRef] [Green Version]
- Guenther, S.; Grobbel, M.; Heidemanns, K.; Schlegel, M.; Ulrich, R.G.; Ewers, C.; Wieler, L.H. First insights into antimicrobial resistance among faecal Escherichia coli isolates from small wild mammals in rural areas. Sci. Total Environ. 2010, 408, 3519–3522. [Google Scholar] [CrossRef]
- Marshall, B.; Petrowski, D.; Levy, S.B. Inter- and intraspecies spread of Escherichia coli in a farm environment in absence of antibiotic usage. Proc. Nat. Acad. Sci. USA 1990, 87, 6609–6613. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Madec, J.Y.; Lupo, A.; Schink, A.K.; Kieffer, N.; Nordmann, P.; Schwarz, S. (Eds.) Antimicrobial Resistance in Escherichia coli. Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; ASM Press: Washington, DC, USA, 2018; pp. 289–316. [Google Scholar]
- Wang, Y.; Zhang, R.; Li, J.; Wu, Z.; Yin, W.; Schwarz, S.; Tyrrell, J.M.; Zheng, Y.; Wang, S.; Shen, Z.; et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017, 2, 16260. [Google Scholar] [CrossRef]
- Hansen, A.K.; Velschow, S. Antibiotic resistance in laboratory animals. Lab. Anim. 2000, 34, 413–422. [Google Scholar]
- Gakuya, F.M.; Kyule, M.N.; Gathura, P.B.; Kariuki, S. Antimicrobial susceptibility and plasmids from Escherichia coli isolated from rats. East Afr. Med J. 2001, 78, 518–522. [Google Scholar] [CrossRef] [Green Version]
- Gakuya, F.M.; Kyule, M.N.; Gathura, P.B.; Kariuki, S. Antimicrobial resistance of bacterial organisms isolated from rats. East Afr. Med J. 2001, 78, 646–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, K.; Blascovich, J.; Mendes, W.B. Cardiovascular reactivity and the presence of pets, friends, and spouses: The truth about cats and dogs. Psychosom. Med. 2002, 64, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Anderson, W.P.; Reid, C.M.; Jennings, G.L. Pet ownership and risk factors for cardiovascular disease. Med J. Aust. 1992, 157, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, E.K.; Nelson, M.R.; Jennings, G.L.; Wing, L.M.; Reid, C.M. Pet ownership and survival in the elderly hypertensive population. J. Hypertens. 2017, 35, 769–775. [Google Scholar] [CrossRef]
- Bryant, B.K. The richness of the child-pet relationship: A consideration of both benefits and costs of pets to children. Anthrozoös 1990, 3, 253–261. [Google Scholar] [CrossRef]
- Poresky, R.H. Companion animals and other factors affecting young children’s development. Anthrozoös 1996, 9, 159–168. [Google Scholar] [CrossRef]
- Poresky, R.H.; Hendrix, C. Differential effects of pet presence and pet-bonding on young children. Psychol. Rep. 1990, 67, 51–54. [Google Scholar] [CrossRef]
- Animal Welfare Institute. Companion Animals. Available online: https://awionline.org/content/companion-animals (accessed on 6 December 2020).
- FEDIAF. The European Pet Food Industry: European Facts and Figures. 2019. Available online: https://www.fediaf.org/images/FEDIAF_facts_and_figs_2019_cor-35-48.pdf (accessed on 7 December 2020).
- Brown, D.C.; Conzemius, M.G.; Shofer, F.; Swann, H. Epidemiologic evaluation of postoperative wound infections in dogs and cats. J. Am. Vet. Med. Assoc. 1997, 210, 1302–1306. [Google Scholar]
- Marques, C.; Belas, A.; Franco, A.; Aboim, C.; Gama, L.T.; Pomba, C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J. Antimicrob. Chemother. 2018, 73, 377–384. [Google Scholar] [CrossRef] [Green Version]
- Moyaert, H.; De Jong, A.; Simjee, S.; Rose, M.; Youala, M.; El Garch, F.; Vila, T.; Klein, U.; Rzewuska, M.; Morrissey, I. Survey of antimicrobial susceptibility of bacterial pathogens isolated from dogs and cats with respiratory tract infections in Europe: ComPath results. J. Appl. Microbiol. 2019, 127, 29–46. [Google Scholar] [CrossRef]
- McMeekin, C.H.; Hill, K.E.; Gibson, I.R.; Bridges, J.P.; Benschop, J. Antimicrobial resistance patterns of bacteria isolated from canine urinary samples submitted to a New Zealand veterinary diagnostic laboratory between 2005–2012. N. Z. Vet. J. 2017, 65, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Penna, B.; Varges, R.; Martins, R.; Martins, G.; Lilenbaum, W. In vitro antimicrobial resistance of staphylococci isolated from canine urinary tract infection. Can. Vet. J. 2010, 51, 738–742. [Google Scholar] [PubMed]
- Cohn, L.A.; Gary, A.T.; Fales, W.H.; Madsen, R.W. Trends in fluoroquinolone resistance of bacteria isolated from canine urinary tracts. J. Vet. Diagn. Investig. 2003, 15, 338–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Liu, Z.; Zhang, Y.; Zhang, Z.; Lei, L.; Xia, Z. Increasing prevalence of ESBL-producing multidrug resistance Escherichia coli from diseased pets in Beijing, China from 2012 to 2017. Front. Microbiol. 2019, 10, 2852. [Google Scholar] [CrossRef] [PubMed]
- De Briyne, N.; Atkinson, J.; Borriello, S.P.; Pokludová, L. Antibiotics used most commonly to treat animals in Europe. Vet. Rec. 2014, 175, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loeffler, A.; Pfeiffer, D.; Lindsay, J.A.; Soares-Magalhaes, R.; Lloyd, D.H. Prevalence of and risk factors for MRSA carriage in companion animals: A survey of dogs, cats and horses. Epidemiol. Infect. 2010, 139, 1019–1028. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, J.; Poeta, P.; Martins, A.; Costa, D. The Importance of pets as reservoirs of resistant enterococcus strains, with special reference to vancomycin. J. Vet. Med. Ser. B 2002, 49, 278–280. [Google Scholar] [CrossRef]
- Iseppi, R.; Di Cerbo, A.; Messi, P.; Sabia, C. Antibiotic resistance and virulence traits in vancomycin-resistant Enterococci (VRE) and extended-spectrum β-lactamase/AmpC-producing (ESBL/AmpC) Enterobacteriaceae from humans and pets. Antibiotics 2020, 9, 152. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, A.; Batabyal, K.; Singh, A.; Joardar, S.; Dey, S.; Isore, D.; Sar, T.; Dutta, T.K.; Bandyopadhayay, S.; Samanta, I. Multi-drug resistant, biofilm-producing high-risk clonal lineage of Klebsiella in companion and household animals. Lett. Appl. Microbiol. 2020, 71, 580–587. [Google Scholar] [CrossRef]
- Loncaric, I.; Rosel, A.C.; Szostak, M.P.; Licka, T.F.; Allerberger, F.; Ruppitsch, W.; Spergser, J. Broad-spectrum cephalosporin-resistant Klebsiella spp. isolated from diseased horses in Austria. Animals 2020, 10, 332. [Google Scholar] [CrossRef] [Green Version]
- Endimiani, A.; Hujer, K.M.; Hujer, A.M.; Bertschy, I.; Rossano, A.; Koch, C.; Gerber, V.; Francey, T.; Bonomo, R.A.; Perreten, V. Acinetobacter baumannii isolates from pets and horses in Switzerland: Molecular characterization and clinical data. J. Antimicrob. Chemother. 2011, 66, 2248–2254. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Haenni, M.; Madec, J.-Y. Antimicrobial resistance in Acinetobacter spp. and Pseudomonas spp. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; Schwartz, S., Cavaco, L.M., Shen, L., Eds.; AMS: Washington, DC, USA, 2018; pp. 377–393. [Google Scholar] [CrossRef]
- Hall, J.L.; Holmes, M.A.; Baines, S.J. Prevalence and antimicrobial resistance of canine urinary tract pathogens. Vet. Rec. 2013, 173, 549. [Google Scholar] [CrossRef]
- Watson, A. Diet and periodontal disease in dogs and cats. Aust. Vet. J. 1994, 71, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Dear, J.D. Bacterial pneumonia in dogs and cats. Vet. Clin. North Am. Small Anim. Pract. 2014, 44, 143–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, R.B. Canine infectious respiratory disease. In Infectious Diseases of the Dog and Cat; Sykes, J., Greene, C.E., Eds.; Saunders: Philadelphia, PA, USA, 2012; p. 55. [Google Scholar]
- Maboni, G.; Seguel, M.; Lorton, A.; Berghaus, R.; Sanchez, S. Canine infectious respiratory disease: New insights into the etiology and epidemiology of associated pathogens. PLoS ONE 2019, 14, e0215817. [Google Scholar] [CrossRef] [Green Version]
- Weese, S.J. Bacterial pneumonia. In Infectious Diseases of the Dog and Cat: A Colour Handbook; Weese, S.J., Evason, M., Eds.; CRC Press: Boca Raton, FL, USA, 2020; pp. 2–3. [Google Scholar]
- Evason, M.; Rutter, C.R. Neurologic diseases. In Infectious Diseases of the Dog and Cat: A Colour Handbook; Weese, S.J., Evason, M., Eds.; CRC Press: Boca Raton, FL, USA, 2020; p. 104. [Google Scholar]
- Evason, M. Respiratory diseases. In Infectious Diseases of the Dog and Cat: A Colour Handbook; Weese, S.J., Evason, M., Eds.; CRC Press: Boca Raton, FL, USA, 2020; p. 16. [Google Scholar]
- Windahl, U.; Holst, B.S.; Nyman, A.-K.J.; Grönlund, U.; Bengtsson, B. Characterisation of bacterial growth and antimicrobial susceptibility patterns in canine urinary tract infections. BMC Vet. Res. 2014, 10, 217. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.-K.; Lo, D.-Y.; Wei, H.-W.; Kuo, H.-C. Antimicrobial resistance of Escherichia coli isolates from canine urinary tract infections. J. Vet. Med. Sci. 2015, 77, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Ghasemzadeh, I.; Namazi, S. Review of bacterial and viral zoonotic infections transmitted by dogs. J. Med. Life 2015, 8, 1–5. [Google Scholar]
- Davies, M.; Stewart, P. Transferable drug resistance in man and animals: Genetic relationship between R-plasmids in enteric bacteria from man and domestic pets. Aust. Vet. J. 1978, 54, 507–512. [Google Scholar] [CrossRef]
- Kühn, I. Epidemiology and ecology of enterococci, with special reference to antibiotic resistant strains, in animals, humans and the environment—Example of an ongoing project within the European research programme. Int. J. Antimicrob. Agents 2000, 14, 337–342. [Google Scholar] [CrossRef]
- De Greeff, S.C.; Mouton, J.W.; Schoffelen, A.F.; Verduin, C.M. NethMap 2019: Consumption of Antimicrobial Agents and Antimicrobial Resistance among Medically Important Bacteria in The Netherlands/MARAN 2019: Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in The Netherlands in 2018; Rijksinstituut voor Volksgezondheid en Milieu RIVM: Utrecht, The Netherlands, 2019. [Google Scholar]
- Schnellmann, C.; Gerber, V.; Rossano, A.; Jaquier, V.; Panchaud, Y.; Doherr, M.; Thomann, A.; Straub, R.; Perreten, V. Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J. Clin. Microbiol. 2006, 44, 4444–4454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares-Magalhaes, R.; Loeffler, A.; Lindsay, J.; Rich, M.; Roberts, L.; Smith, H.; Lloyd, D.H.; Pfeiffer, D.U. Risk factors for methicillin-resistant Staphylococcus aureus (MRSA) infection in dogs and cats: A case-control study. Vet. Res. 2010, 41, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, I.; Pimenta, P.; Cunha, R.; Martins, C.; Silva, V.; Igrejas, G.; Torres, C.; Poeta, P. Cefotaxime, Imipenem and Colistin resistance in Klebsiella pneumoniae from pets in Portugal. In Proceedings of the Microbiotech ’17, Porto, Portugal, 7–9 December 2017; Available online: https://www.researchgate.net/profile/Isabel_Carvalho11/publication/332963802_Cefotaxime_Imipenem_and_Colistin_resistance_in_Klebsiella_pneumoniae_from_pets_in_Portugal/links/5cd59e36a6fdccc9dd9ee19c/Cefotaxime-Imipenem-and-Colistin-resistance-in-Klebsiella-pneumoniae-from-pets-in-Portugal.pdf (accessed on 5 December 2020).
- Chen, J.-W.; Huang, H.H.; Chang, S.-M.; Scaria, J.; Chiu, Y.-L.; Chen, C.-M.; Ko, W.-C.; Hung, C.-H. Antibiotic-resistant Escherichia coli and sequence type 131 in fecal colonization in dogs in Taiwan. Microorganisms 2020, 8, 1439. [Google Scholar] [CrossRef] [PubMed]
- Hartantyo, S.H.P.; Chau, M.L.; Fillon, L.; Ariff, A.Z.B.M.; Kang, J.S.L.; Aung, K.T.; Gutiérrez, R.A. Sick pets as potential reservoirs of antibiotic-resistant bacteria in Singapore. Antimicrob. Resist. Infect. Control. 2018, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.A.; Whitlock, J.E.; Harwood, V.J. Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl. Environ. Microbiol. 2006, 72, 6914–6922. [Google Scholar] [CrossRef] [Green Version]
- Woolhouse, M.E.J.; Waugh, C.; Perry, M.R.; Nair, H. Global disease burden due to antibiotic resistance—State of the evidence. J. Glob. Health 2016, 6, 010306. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [Green Version]
- Martínez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; De Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial resistance in the food chain: A Review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef] [Green Version]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.H.M.; Findlay, D.; Gyssens, I.C.J.; Heure, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Dupouy, V.; Abdelli, M.; Moyano, G.; Arpaillange, N.; Bibbal, D.; Cadiergues, M.-C.; Lopez-Pulin, D.; Sayah-Jeanne, S.; De Gunzburg, J.; Saint-Lu, N.; et al. Prevalence of beta-lactam and quinolone/fluoroquinolone resistance in Enterobacteriaceae from dogs in France and Spain—Characterization of ESBL/pAmpC isolates, genes, and conjugative plasmids. Front. Vet. Sci. 2019, 6, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ljungquist, O.; Ljungquist, D.; Myrenås, M.; Rydén, C.; Finn, M.; Bengtsson, B. Evidence of household transfer of ESBL-/pAmpC-producing Enterobacteriaceae between humans and dogs—A pilot study. Infect. Ecol. Epidemiol. 2016, 6, 31514. [Google Scholar] [CrossRef] [PubMed]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watkins, R.R.; Bonomo, R.A. Overview: Global and local impact of antibiotic resistance. Infect. Dis. Clin. N. Am. 2016, 30, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Gwanzura, L.; Pasi, C.; Nathoo, K.J.; Hakim, J.; Gangaidzo, I.; Mielke, J.; Robertson, V.J.; Heyderman, R.S.; Mason, P.R. Rapid emergence of resistance to penicillin and trimethoprim–sulphamethoxazole in invasive Streptococcus pneumoniae in Zimbabwe. Int. J. Antimicrob. Agents 2003, 21, 557–561. [Google Scholar] [CrossRef]
- Aworh, M.K.; Kwaga, J.; Okolocha, E.; Mba, N.; Thakur, S. Prevalence and risk factors for multi-drug resistant Escherichia coli among poultry workers in the Federal Capital Territory, Abuja, Nigeria. PLoS ONE 2019, 14, e0225379. [Google Scholar] [CrossRef]
- Cho, S.-H.; Lim, Y.-S.; Kang, Y.-H. Comparison of antimicrobial resistance in Escherichia coli strains isolated from healthy poultry and swine farm workers using antibiotics in Korea. Osong Public Health Res. Perspect. 2012, 3, 151–155. [Google Scholar] [CrossRef] [Green Version]
- Saengthongpinit, C.; Sratonhno, K.; Phimpraphai, P.T.; Morand, S.; de Garine-Wichatitsky, M. Antimicrobial resistance of Salmonella spp. isolates and heavy metal traces from rodent meat purchased from roadside markets, Central Thailand. Pathog. Dis. 2019, 16, 687–695. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Samanta, I. Antimicrobial Resistance in agri-food chain and companion animals as a re-emerging menace in post-COVID epoch: Low-and middle-income countries perspective and mitigation strategies. Front. Vet. Sci. 2020, 7, 620. [Google Scholar] [CrossRef]
- Friant, S.; Ayambem, W.A.; Alobi, A.O.; Ifebueme, N.M.; Otukpa, O.M.; Ogar, D.A.; Alawa, C.B.; Goldberg, T.L.; Jacka, J.K.; Rothman, J.M. Eating bushmeat improves food security in a biodiversity and infectious disease “hotspot”. EcoHealth 2020, 17, 1–14. [Google Scholar] [CrossRef]
- Naseem, R.; Majeed, W.; Rana, N.; Koch, E.B.; Naseem, M.R. Entomophagy: An innovative nutritional and economic navigational tool in race of food security. Int. J. Trop. Insect Sci. 2020, 1–11. [Google Scholar] [CrossRef]
- Mahamat, O.O.; Kempf, M.; Lounnas, M.; Tidjani, A.; Hide, M.; Benavides, J.; Carrière, C.; Bañuls, A.L.; Jean-Pierre, H.; Ouedraogo, A.S.; et al. Epidemiology and prevalence of extended-spectrum β-lactamase-and carbapenemase-producing Enterobacteriaceae in humans, animals and environments in West and Central Africa. Int. J. Antimicrob. Agents 2020, 57, 106203. [Google Scholar] [CrossRef]
- Gwenzi, W.; Chaukura, N. Organic contaminants in African aquatic systems: Current knowledge, health risks, and future research directions. Sci. Total Environ. 2018, 619, 1493–1514. [Google Scholar] [CrossRef] [PubMed]
- Faleye, A.; Adegoke, A.; Ramluckan, K.; Bux, F.; Stenström, T.A. Antibiotic residue in the aquatic environment: Status in Africa. Open Chem. 2018, 16, 890–903. [Google Scholar] [CrossRef]
- Loewenson, R. Occupational health epidemiology in Africa: Drought on fertile grounds. Int. J. Occup. Environ. Health 1995, 1, 260–268. [Google Scholar] [CrossRef]
- Pepper, I.L.; Brooks, J.P.; Gerba, C.P. Antibiotic resistant bacteria in municipal wastes: Is there reason for concern? Environ. Sci. Technol. 2018, 52, 3949–3959. [Google Scholar] [CrossRef]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar] [CrossRef]
- Claycamp, H.G. Chapter 14—Risk assessment of antimicrobial resistance. In Antimicrobial Resistance and Food Safety; Chen, C.-Y., Yan, X., Jackson, C.R., Eds.; Academic Press: San Diego, CA, USA, 2015; pp. 283–302. [Google Scholar]
- Ashbolt, N.J.; Amézquita, A.; Backhaus, T.; Borriello, P.; Brandt, K.K.; Collignon, P.; Coors, A.; Finley, R.; Gaze, W.H.; Heberer, T.; et al. Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013, 121, 993–1001. [Google Scholar] [CrossRef] [Green Version]
- European Medicines Agency. Reflection Paper on the Risk of Antimicrobial Resistance Transfer from Companion Animals; European Medicines Agency: London, UK, 2015. [Google Scholar]
- World Health Organization. WHO List of Critically Important Antimicrobials for Human Medicine (WHO CIA List); World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Zhang, X.-F.; Doi, Y.; Huang, X.; Li, H.-Y.; Zhong, L.-L.; Zeng, K.-J.; Zhang, Y.-F.; Patil, S.; Tian, G.-B. Possible transmission of mcr-1-harboring Escherichia coli between companion animals and human. Emerg. Infect. Dis. 2016, 22, 1679–1681. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, A.; Barbosa, A.; Arais, L.; Ribeiro, P.; Carneiro, V.; Cerqueira, A. Resistance patterns, ESBL genes, and genetic relatedness of Escherichia coli from dogs and owners. Braz. J. Microbiol. 2016, 47, 150–158. [Google Scholar] [CrossRef] [Green Version]
- Damborg, P.; Nielsen, S.S.; Guardabassi, L. Escherichia coli shedding patterns in humans and dogs: Insights into within-household transmission of phylotypes associated with urinary tract infections. Epidemiol. Infect. 2009, 137, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Loeber, M.; Jacobson, A. Transmission of multiple antimicrobial-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet. Microbiol. 2004, 98, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Founou, R.C.; Founou, L.L.; Essack, S.Y. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0189621. [Google Scholar] [CrossRef] [Green Version]
- Sifri, Z.C.; Chokshi, A.; Cennimo, D.; Horng, H. Global contributors to antibiotic resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [CrossRef]
- Childs, J.E.; Cheek, J.E.; Rollin, P.E.; Enscore, R.E.; Maupin, G.O.; Glass, G.E.; Zeitz, P.S.; Krebs, J.W.; Butler, J.C.; Gage, K.L.; et al. A household-based, case-control study of environmental factors associated with hantavirus pulmonary syndrome in the southwestern United States. Am. J. Trop. Med. Hyg. 1995, 52, 393–397. [Google Scholar] [CrossRef]
- Costa, F.; Ribeiro, G.S.; Felzemburgh, R.D.M.; Santos, N.; Reis, R.B.; Santos, A.C.; Fraga, D.B.M.; Araujo, W.N.; Santana, C.; Childs, J.E.; et al. Influence of household rat infestation on Leptospira transmission in the urban slum environment. PLoS Neglected Trop. Dis. 2014, 8, e3338. [Google Scholar] [CrossRef] [Green Version]
- Munoz-Zanzi, C.; Mason, M.; Encina, C.; Gonzalez, M.; Berg, S. Household characteristics associated with rodent presence and Leptospira infection in rural and urban communities from southern Chile. Am. J. Trop. Med. Hyg. 2014, 90, 497–506. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Antimicrobial Resistance; Fact Sheet No. 194; WHO: Geneva, Switzerland, 2013. [Google Scholar]
- Kumsa, B.; Socolovschi, C.; Parola, P.; Rolain, J.-M.; Raoult, D. molecular detection of Acinetobacter species in lice and keds of domestic animals in Oromia Regional State, Ethiopia. PLoS ONE 2012, 7, e52377. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Macovei, L.; Miles, B.; Zurek, L. Potential of houseflies to contaminate ready-to-eat food with antibiotic-resistant enterococci. J. Food Prot. 2008, 71, 435–439. [Google Scholar] [CrossRef]
- Roncolini, A.; Cardinali, F.; Aquilanti, L.; Milanović, V.; Garofalo, C.; Sabbatini, R.; Abaker, M.S.S.; Pandolfi, M.; Pasquini, M.; Tavoletti, S.; et al. Investigating antibiotic resistance genes in marketed ready-to-eat small crickets (Acheta domesticus). J. Food Sci. 2019, 84, 3222–3232. [Google Scholar] [CrossRef] [PubMed]
- Di Liberto, D.; Locati, M.; Caccamo, N.; Vecchi, A.; Meraviglia, S.; Salerno, A.; Sireci, G.; Nebuloni, M.; Caceres, N.; Cardona, P.-J.; et al. Role of the chemokine decoy receptor D6 in balancing inflammation, immune activation, and antimicrobial resistance in Mycobacterium tuberculosis infection. J. Exp. Med. 2008, 205, 2075–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karam, M.R.A.; Habibi, M.; Bouzari, S. Relationships between virulence factors and antimicrobial resistance among Escherichia coli Isolated from urinary tract infections and commensal isolates in Tehran, Iran. Osong Public Health Res. Perspect. 2018, 9, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Cook, L.C.C.; Shu, C.-C.; Chen, Y.; Manias, D.A.; Ramkrishna, D.; Dunny, G.M.; Hu, W.-S. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc. Natl. Acad. Sci. USA 2013, 110, 7086–7090. [Google Scholar] [CrossRef] [Green Version]
- Kalsy, M.; Tonk, M.; Hardt, M.; Dobrindt, U.; Zdybicka-Barabas, A.; Cytrynska, M.; Vilcinskas, A.; Mukherjee, K. The insect antimicrobial peptide cecropin A disrupts uropathogenic Escherichia coli biofilms. NPJ Biofilms Microbiomes 2020, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Moretta, A.; Salvia, R.; Scieuzo, C.; Di Somma, A.; Vogel, H.; Pucci, P.; Sgambato, A.; Wolff, M.; Falabella, P. A bioinformatic study of antimicrobial peptides identified in the Black Soldier Fly (BSF) Hermetia illucens (Diptera: Stratiomyidae). Sci. Rep. 2020, 10, 16875. [Google Scholar] [CrossRef]
- Dallavecchia, D.L.; Ricardo, E.; Da Silva, A.S.; Rodrigues, A.G. Antibacterial and antifungal activity of excretions and secretions of Calliphora vicina. Med Vet. Entomol. 2020. [Google Scholar] [CrossRef]
- Feng, M.; Fei, S.; Xia, J.; Labropoulou, V.; Swevers, L.; Sun, J. Antimicrobial peptides as potential antiviral factors in insect antiviral immune response. Front. Immunol. 2020, 11, 2030. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Prasad, A.S.B.; Mehta, C.H.; Nayak, U.Y. Antimicrobial peptide polymers: No escape to ESKAPE pathogens—A review. World J. Microbiol. Biotechnol. 2020, 36, 1–14. [Google Scholar] [CrossRef]
- Hawkey, K.J.; Lopez-Viso, C.; Brameld, J.M.; Parr, T.; Salter, A.M. Insects: A potential source of protein and other nutrients for feed and food. Annu. Rev. Anim. Biosci. 2021, 9. [Google Scholar] [CrossRef]
- Nyberg, M.; Olsson, V.; Wendin, K. Reasons for eating insects? Responses and reflections among Swedish consumers. Int. J. Gastron. Food Sci. 2020, 22, 100268. [Google Scholar] [CrossRef]
- Liu, C.; Yao, H.; Chapman, S.J.; Su, J.; Wang, C. Changes in gut bacterial communities and the incidence of antibiotic resistance genes during degradation of antibiotics by black soldier fly larvae. Environ. Int. 2020, 142, 105834. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Ibrahim, M.; Ahmad, F.; Rana, H.A.; Rao, T.; Anwar, W.; Younus, M.; Ahmad, W.; Farooqi, S.H.; Aftab, A. Microbial risk assessment and antimicrobial resistance. In Antibiotics and Antimicrobial Resistance Genes in the Environment; Hashmi, M.Z., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 313–330. [Google Scholar]
- Valli, R.X.E.; Lyng, M.; Kirkpatrick, C.L. There is no hiding if you Seq: Recent breakthroughs in Pseudomonas aeruginosa research revealed by genomic and transcriptomic next-generation sequencing. J. Med Microbiol. 2020, 69, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.M.; Eng, W.W.H.; Dhanoa, A. First genomic insights into carbapenem-resistant Klebsiella pneumoniae from Malaysia. J. Glob. Antimicrob. Resist. 2020, 20, 153–159. [Google Scholar] [CrossRef]
| Insect Name | AMR Organism | AMR Type | ARG Detected | Reference |
|---|---|---|---|---|
| Houseflies (Musca domestica) Cockroaches (Blattaria/Blattodea) | Enterococci (E. casseliflavus; E. hirae; E. faecium; E. faecalis) | Tetracycline, Erythromycin | Tetracycline tet(M) Erythromycin erm(B) Tn916/1545 transposon family, gelatinase gelE, esp, and asa1 | [37] |
| tet(M) and erm(B) and Tn916/1545 transposon family | ||||
| Cockroaches (Periplaneta Americana Blattella germanica) | 38 species of gram-negative bacteria, 20 species of glucose non-fermenter bacilli and 6 species of gram-positive bacteria | Ampicillin; Gentamicin; Ciprofloxacin, Ofloxacin; Chloramphenicol Tetracycline; Trimethoprim-sulfamethoxazole; Penicillin; Streptomycin; Erythromycin; Oxacillin; Vancomycin; Cephalothin; Ceftazidime; Imipenem; Piperacillin; Cefoperazone | Not determined | [43] |
| Houseflies (Musca domestica) | E. faecalis | Tetracycline; Erythromycin; Streptomycin; Ciprofloxacin; Kanamycin | transposon Tn916 and members of the Tn916/Tn1545 family | [36] |
| Houseflies (Musca domestica) | E. coli | Tetracycline; Ampicillin; Streptomycin; Sulfonamides; Trimethoprim-Sulfamethoxazole; Chloramphenicol; Nalidixic acid | Tetracycline tetA and tetB, sulphonamide sul1, sul2, sul3, extended-spectrum b-lactamase blaTEM, strA | [142] |
| Stored-product grain insects, e.g., darkling beetle, A. diaperinus; Lesser grain borer, Rhyzopertha dominica; Foreign grain beetle, Ahasverus advena; red flour beetle, Tribolium castaneum (Herbst); warehouse beetle, Trogoderma variabile Ballion; | E. casseliflavus; E. gallinarum; E. faecium; E. faecalis; E. hirae | Tetracycline; Streptomycin; Erythromycin; Kanamycin; Ciprofloxacin; Ampicillin; Chloramphenicol | gelatinase gelE; enterococcal surface protein esp; cytolysin cylA | [143,144] |
| Ants (Tapinoma melanocephalum (Fabricius) and Camponotus vittatus (Forel) | Coagulase-positive Staphylococcus (S. aureus); Coagulase-negative Staphylococcus; Gram negative Bacilli | Cephalotine; Oxacillin; Penicillin; Tetracycline; Vancomycin; Ampycillin; Cephalotine; Ciprofloxacine; Sulphazotrin | Not determined | [145] |
| Gypsy moth larvae (Lymantria dispar L.) | Enterococcus spp.; members of the Enterobacteriaceae | Carbenicillin; Ceftazidime; Gentamicin; Erythromycin; Kanamycin; Streptomycin; Vancomycin; Chloramphenicol; Rifampin; Nalidixic acid; Tetracycline. | ramA; sdeX; sdeY; blaLRG-1 | [146] |
| Bedbugs (Cimex lectularius L.) | E. faecium, S. aureus | Vancomycin; Methicillin; Ampicillin; Teicoplanin; Aminoglycosides Erythromycin | Not determined | [147] |
| Honeybees (Apis mellifera L.) | Snodgrassella alvi (Betaproteobacteria) Alphaproteobacteria | Tetracycline Oxytetracyline | tetB, tetC, tetD, tetH, tetL, tetY tetM tetW | [81] |
| Houseflies (Musca domestica) False stable flies (Muscina stabulans) | E. coli | Tetracycline; Streptomycin; Kanamycin; Ampicillin; Cefazolin; Cefpodoxime; Trimethoprim | extended-spectrum b-lactamase (ESBL) blaCTX-M-15 | [65] |
| Flea midgut (Xenopsylla cheopis) | Yersinia pestis | Streptomycin; Gentamycin; Tetracycline; Chloramphenicol; Sulphonamides | Not determined | [111] |
| Houseflies (Musca domestica) blow-flies (Lucilicia species) and Bottle flies (Phaenicia species) | Enterococci (E. casseliflavus; E. gallinarum; E. faecium; E. faecalis) and Staphylococci (S. saprophyticus; S. aureus; S. xylosus; S. epidermidis) | Penicillin, Quinupristin-dalfopristin; Erythromycin; Tetracycline; Clindamycin | erm(B); erm(A); msr(C); msr(A/B); transposon Tn916 | [66] |
| Rodent Species | Microbial Species | AMR Profile | Comments | Reference |
|---|---|---|---|---|
| Rat species 1 | S. aureus, E. coli Pasteurella pnemutropica | Amp (75), Pen (75), AM/Cl (75), Te (12.5) Amp (11.1), Pen (100), Te (25) Amp (11.1), Pen (33.3), AM/Cl (22.2), Te (11.1) | Number in brackets equals % resistance | [176] |
| Mice species 1 | S. aureus, E. coli P. pnemutropica | Amp (87.5), Pen (8.5), AM/Cl (37.5), Te (12.5) Pen (8.5) Pen (20) | Number in brackets equals % resistance | [176] |
| Rat species 2 | E. coli | Amp (23.3), Strep (15), Tmc (6.6), Te (3.3), Amc (1.7) | Number in brackets equals % resistance | [177] |
| Apodemus sylvaticus | E. coli | Amp (48) (21) Tm (33) (8) Cf (21) (4) Ctx (18) (0) | No//in brackets indicate no//of animals with resistant E. coli over no//of animals trapped in coastal vs. inland habitats | [36] |
| Rat species 3 | E. coli K. pneumonia Pseudomonus paucimobilis Chryseomonas luteola Aeromonas caviae Burkhoddria cepacia | Te (16.7), Gn (16.7), Apr (16.7), S3 (66.6), C (83.3), Crd (66.6), Cxm (33.3), Amp (50), Na (50), Tm (50) S3 (50), C (100), Crd (100), Amc (100), Cxm (50), Amp (100), Tm (100) Te (12.9), S3 (9.7), C (12.9), Ctx (67.7), Crd (3.2), Amc (9.7), Cxm (16.1), Amp (6.5), Na (41.9), Tm (6.5) Te (15.4), Gn (7.7), Apr (7.7), S3 (23.1), Ctx (92.3), Crd (53.8), Amc (46.2), Cxm (53.8), Amp (4.2), Na (77), Tm (46.2) Te (37.5), Gn (25), S3 (50), Ctx (50), Crd (25), Amc (37.5), Cxm (62.5), Amp (62.5), Na (87.5), Tm (87.5) Te (66.6), Gn (66.6), S3 (66.6), Ctx (66.6), Amc (16.7), Cxm (83.3), Amp (66.6), Na (100), Tm (66.6) | Number in brackets equals % resistance | [178] |
| Wild rodents | Hafnia alvei E. coli Serratia liquefaciens | Te (76), Tm (10), Na (24), Ac (98), Ap (95), Cxm (100) Te (14), Na (9), Ac (97), Ap (89), Cxm (100) Te (63), Tm (30), Na (30), Ac (100), Ap (97), Cxm (90) | Number in brackets equals % resistance | [159] |
| Wild rodents | Alcaligenes spp. Serratia fonticola Enterobacter intermedius Enterobacter amnigenus Cedacae davisiae Providencia rustigianii | Te (44), Tm (67), Na (56), Ac (67), Ap (67), Cxm (78) Te (50), Na (22), Ac (72), Ap (94), Cxm (67) Te (39), Tm (23), Na (23) Ac (85), Ap (92), Cxm (77) Te (50), Na (40), Ac (90), Ap (100), Cxm (90) Te (44), Tm (22), Na (22), Ac (67), Ap (89), Cxm (89) Te (17), Tm (17), Na (17), Ac (100), Ap (83), Cxm (67) | Number in brackets equals % resistance | [159] |
| Rattus rattus, Rattus norvegicus, Mus musculus | E. coli | Amp (35), Te (15), Ap (10), Na (10), C (5), S3 (5), Cf (5), Nf (5) | Number in brackets equals % of isolates showing resistant phenotype | [161] |
| Pet | Disease | Causative Bacteria | Reference |
|---|---|---|---|
| Dogs | Urinary tract infections, | E. coli, Staphylococcus spp. (S. intermedius, S. aureus), P. aeruginosa, E. faecalis, Proteus mirabilis | [202] |
| Dental | Actinom-vces spp., Streptococcus spp. and other species | [203] | |
| Canine infectious respiratory disease (Kennel Cough) | several viruses/bacteria Streptococcus equi subsp. zooepidemicus Bordetella bronchiseptica Mycoplasma ureaplasma, Mycoplasma acholeplasma, Mycoplasma cynos | [204,205,206] | |
| Pasteurellosis | Pasteurella spp. | ||
| Bacterial pneumonia | Enterobacteriaceae (e.g., E. coli, Enterobacter spp., Klebsiella spp.), Pasteurella spp., Bordetella bronchiseptica, Streptococcus spp. Clostridium tetani | [204,207] | |
| Tetanus (cats) | [208] | ||
| Cats Horses | Urinary tract infections Chlamydiosis Tetanus (cats) Strangles Botulism Tetanus (Lockjaw) | Chlamedia felis C. tetani Streptococcus equi Clostridium botulinum C. tetani | [208,209] |
| Pet | Resistance | Bacteria | Country | Reference |
|---|---|---|---|---|
| Dogs, Cats | Ampicillin (18%) | E. coli | Belgium, Italy, Netherlands | [24] |
| Dogs | Imipenem (0.2%), colistin (5%) | K. pneumoniae | Portugal | [218] |
| Methicillin | S. aureus | [209] | ||
| Dogs, cats | Cefotaxime (66–75%) Ceftazidime (71–80%) | E. coli | Italy | [197] |
| Dogs | Cefazolin (43%), fluoroquinolone (22%) | E. coli | Taiwan | [219] |
| Dogs, Cats | chloramphenicol, tetracycline, doxycycline, co-trimoxazole, ampicillin, cefotaxime | Klebsiella spp. | India | [198] |
| amoxicillin (4.8%), ampicillin (21.2%), lincomycin (98%), tetracycline (95%)_enrofloxacine(77%), ofloxacin (64%), ciprofloxacin (73%) | Enterococcus spp. (E. faecium, E. avium, E. faecalis) | Portugal | [196] | |
| Dogs, Cats | Methicillin | Staphylococcus pseudintermedius (63%), S. aureus (50%) | Singapore | [220] |
| Fluoroquinolone | E. coli (40%) | [220] | ||
| Carbapenem | K. pneumoniae (7%) | [220] | ||
| Cats | Ampicillin (42%), amoxicillin/clavulanate (53%), erythromycin 40%), tetracycline (24%), ciprofloxacin (63%), teicoplanin, vancomycin (24%) | E. faecium, E. faecalis | Italy | [197] |
| Horses | Cephalosporin, ciprofloxacin Gentamycin, tobramycin | K. pneumoniae | Austria | [199] |
| Horses | Several antibiotics | E. coli | USA | [221] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gwenzi, W.; Chaukura, N.; Muisa-Zikali, N.; Teta, C.; Musvuugwa, T.; Rzymski, P.; Abia, A.L.K. Insects, Rodents, and Pets as Reservoirs, Vectors, and Sentinels of Antimicrobial Resistance. Antibiotics 2021, 10, 68. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10010068
Gwenzi W, Chaukura N, Muisa-Zikali N, Teta C, Musvuugwa T, Rzymski P, Abia ALK. Insects, Rodents, and Pets as Reservoirs, Vectors, and Sentinels of Antimicrobial Resistance. Antibiotics. 2021; 10(1):68. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10010068
Chicago/Turabian StyleGwenzi, Willis, Nhamo Chaukura, Norah Muisa-Zikali, Charles Teta, Tendai Musvuugwa, Piotr Rzymski, and Akebe Luther King Abia. 2021. "Insects, Rodents, and Pets as Reservoirs, Vectors, and Sentinels of Antimicrobial Resistance" Antibiotics 10, no. 1: 68. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10010068








