Genome-Based Analyses of Fitness Effects and Compensatory Changes Associated with Acquisition of blaCMY-, blaCTX-M-, and blaOXA-48/VIM-1-Containing Plasmids in Escherichia coli
Abstract
:1. Introduction
2. Results
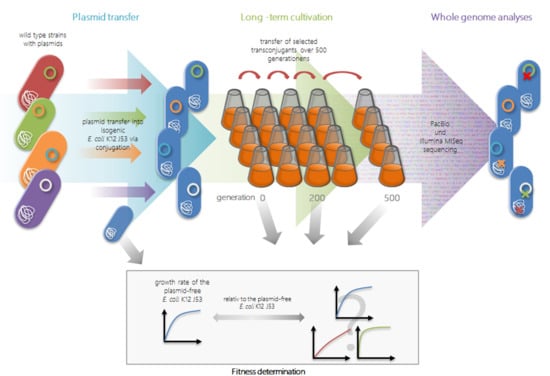
2.1. Transfer Experiments
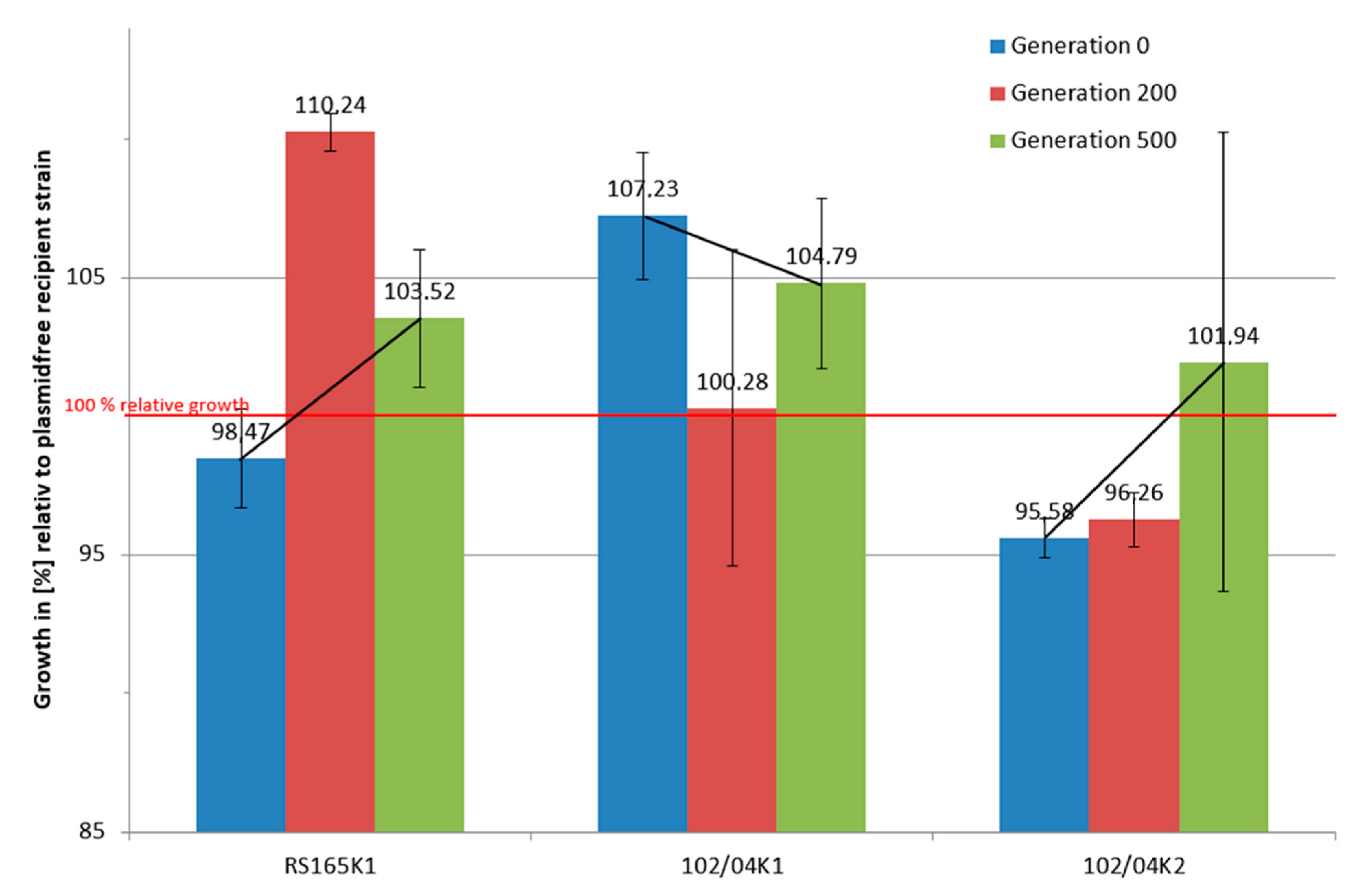
2.2. Long-Term Growth Experiments
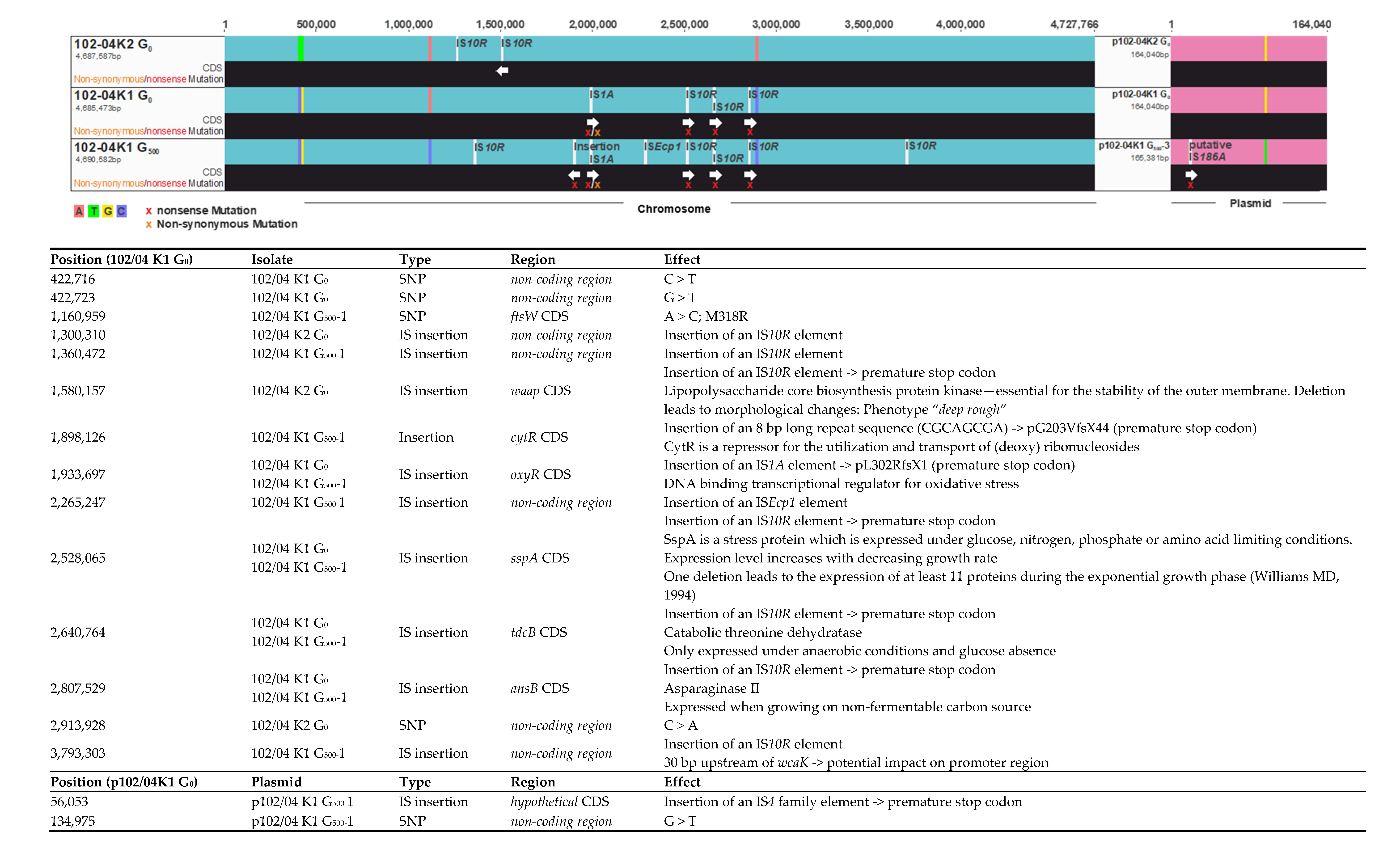
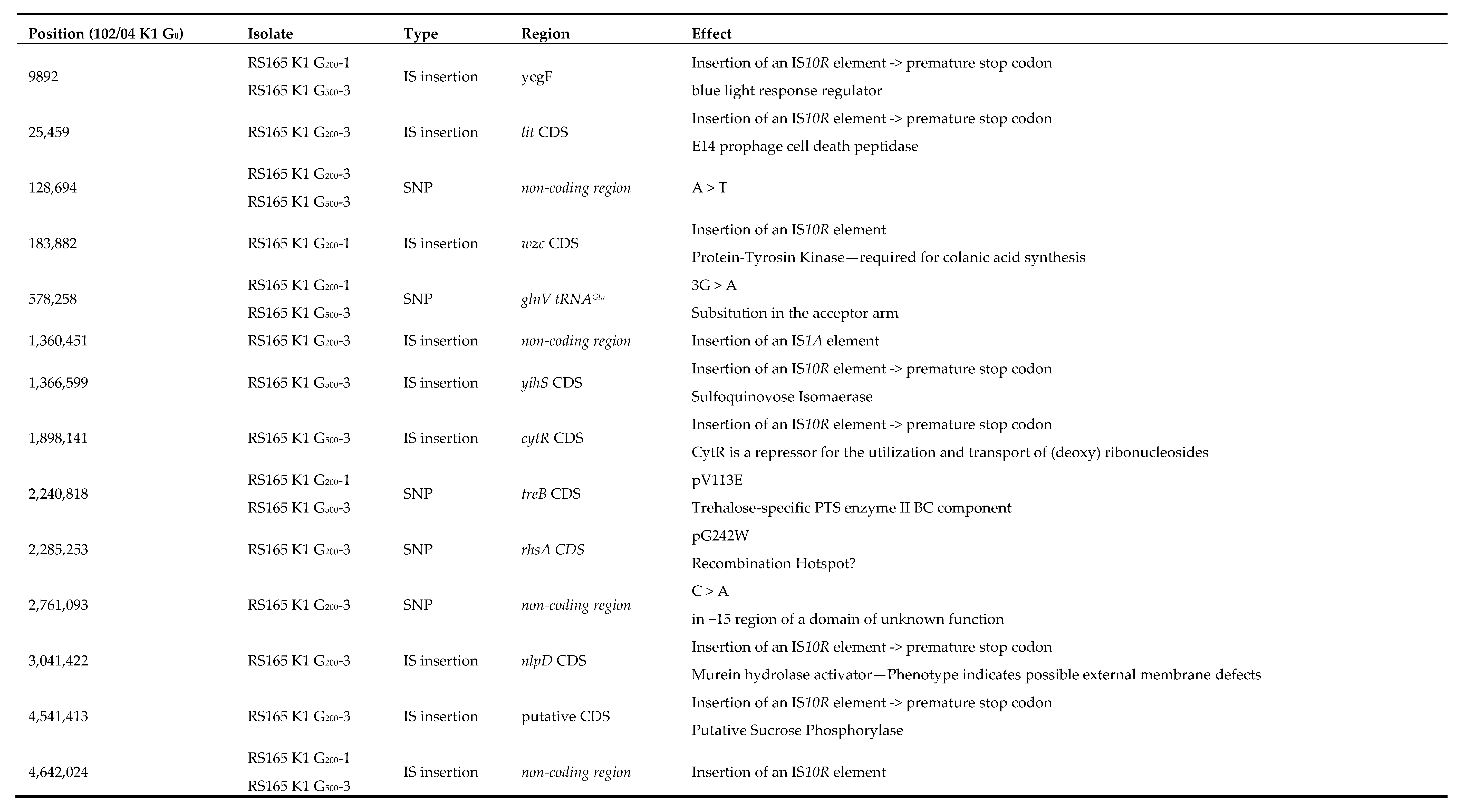
2.3. Genome Reconstruction and Genome Comparisons
3. Discussion
3.1. Comparative Growth Experiments
3.2. Genome Comparisons
3.3. Long-Term Growth Analysis
3.4. Limitations of the Study
4. Materials and Methods
4.1. Strains
4.2. Transfer Experiments and Characterization of Transconjugants
4.3. Growth Experiments in Microtiter Plates
4.4. Long-Term Growth Experiments
4.5. Determining Growth Characteristics and Relative Fitness
4.6. Isolation of DNA, Plasmid DNA and Genomic DNA for Sequencing
4.7. DNA Sequencing by NGS and SMRT Sequencing
4.8. Whole-Genome Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA(EARS-Net)-Annual Epidemiological Report 2019; European Centre for Disease Prevention and Control: Stockholm, Sweden, 18 November 2020. [Google Scholar]
- Praski Alzrigat, L.; Huseby, D.L.; Brandis, G.; Hughes, D. Fitness cost constrains the spectrum of marR mutations in ciprofloxacin-resistant Escherichia coli. J. Antimicrob. Chemother. 2017, 72, 3016–3024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinel, C.; Cacaci, M.; Meignen, P.; Guérin, F.; Davies, B.W.; Sanguinetti, M.; Giard, J.-C.; Cattoir, V. Subinhibitory Concentrations of Ciprofloxacin Enhance Antimicrobial Resistance and Pathogenicity of Enterococcus faecium. Antimicrob. Agents Chemother. 2017, 61, e02763-16. [Google Scholar] [CrossRef] [Green Version]
- Melnyk, A.H.; Wong, A.; Kassen, R. The fitness costs of antibiotic resistance mutations. Evol. Appl. 2015, 8, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Freihofer, P.; Akbergenov, R.; Teo, Y.; Juskeviciene, R.; Andersson, D.I.; Bottger, E.C. Nonmutational compensation of the fitness cost of antibiotic resistance in mycobacteria by overexpression of tlyA rRNA methylase. RNA 2016, 22, 1836–1843. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.; Brandis, G. Rifampicin Resistance: Fitness Costs and the Significance of Compensatory Evolution. Antibiotics 2013, 2, 206–216. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, K.L.; Pedersen, T.M.; Udekwu, K.I.; Petersen, A.; Skov, R.L.; Hansen, L.H.; Hughes, D.; Frimodt-Møller, N. Fitness cost: A bacteriological explanation for the demise of the first international methicillin-resistant Staphylococcus aureus epidemic. J. Antimicrob. Chemother. 2012, 67, 1325–1332. [Google Scholar] [CrossRef]
- Knoppel, A.; Nasvall, J.; Andersson, D.I. Compensating the Fitness Costs of Synonymous Mutations. Mol. Biol. Evol. 2016, 33, 1461–1477. [Google Scholar] [CrossRef] [Green Version]
- Knoppel, A.; Lind, P.A.; Lustig, U.; Nasvall, J.; Andersson, D.I. Minor fitness costs in an experimental model of horizontal gene transfer in bacteria. Mol. Biol. Evol. 2014, 31, 1220–1227. [Google Scholar] [CrossRef] [Green Version]
- Baltrus, D.A. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 2013, 28, 489–495. [Google Scholar] [CrossRef]
- Botelho, J.; Schulenburg, H. The Role of Integrative and Conjugative Elements in Antibiotic Resistance Evolution. Trends Microbiol. 2021, 29, 8–18. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef] [PubMed]
- Meini, S.; Tascini, C.; Cei, M.; Sozio, E.; Rossolini, G.M. AmpC β-lactamase-producing Enterobacterales: What a clinician should know. Infection 2019, 47, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Doi, Y.; Bonomo, R.A.; Johnson, J.K.; Simner, P.J. A Primer on AmpC β-Lactamases: Necessary Knowledge for an Increasingly Multidrug-resistant World. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2019, 69, 1446–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathers, A.J.; Peirano, G.; Pitout, J.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seiffert, S.N.; Hilty, M.; Perreten, V.; Endimiani, A. Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: An emerging problem for human health? Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2013, 16, 22–45. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves Ribeiro, T.; Novais, Â.; Machado, E.; Peixe, L. Acquired AmpC β-Lactamases among Enterobacteriaceae from Healthy Humans and Animals, Food, Aquatic and Trout Aquaculture Environments in Portugal. Pathogens 2020, 9, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, P.N.A.; Ben Zakour, N.L.; Roberts, L.W.; Wailan, A.M.; Zowawi, H.M.; Tambyah, P.A.; Lye, D.C.; Jureen, R.; Lee, T.H.; Yin, M.; et al. Whole genome analysis of cephalosporin-resistant Escherichia coli from bloodstream infections in Australia, New Zealand and Singapore: High prevalence of CMY-2 producers and ST131 carrying blaCTX-M-15 and blaCTX-M-27. J. Antimicrob. Chemother. 2018, 73, 634–642. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Imirzalioglu, C.; Ghosh, H.; Gwozdzinski, K.; Schmiedel, J.; Gentil, K.; Bauerfeind, R.; Kämpfer, P.; Seifert, H.; Michael, G.B.; et al. Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int. J. Antimicrob. Agents 2016, 47, 457–465. [Google Scholar] [CrossRef]
- Pietsch, M.; Eller, C.; Wendt, C.; Holfelder, M.; Falgenhauer, L.; Fruth, A.; Grössl, T.; Leistner, R.; Valenza, G.; Werner, G.; et al. Molecular characterisation of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates from hospital and ambulatory patients in Germany. Vet. Microbiol. 2017, 200, 130–137. [Google Scholar] [CrossRef]
- Valentin, L.; Sharp, H.; Hille, K.; Seibt, U.; Fischer, J.; Pfeifer, Y.; Michael, G.B.; Nickel, S.; Schmiedel, J.; Falgenhauer, L.; et al. Subgrouping of ESBL-producing Escherichia coli from animal and human sources: An approach to quantify the distribution of ESBL types between different reservoirs. Int. J. Med. Microbiol. IJMM 2014, 304, 805–816. [Google Scholar] [CrossRef] [Green Version]
- Denkel, L.A.; Maechler, F.; Schwab, F.; Kola, A.; Weber, A.; Gastmeier, P.; Pfäfflin, F.; Weber, S.; Werner, G.; Pfeifer, Y.; et al. Infections caused by extended-spectrum β-lactamase-producing Enterobacterales after rectal colonization with ESBL-producing Escherichia coli or Klebsiella pneumoniae. Clin. Microbiol. Infect. 2019. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, M.; Irrgang, A.; Roschanski, N.; Brenner Michael, G.; Hamprecht, A.; Rieber, H.; Käsbohrer, A.; Schwarz, S.; Rösler, U.; Kreienbrock, L.; et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef] [PubMed]
- Billard-Pomares, T.; Clermont, O.; Castellanos, M.; Magdoud, F.; Royer, G.; Condamine, B.; Fouteau, S.; Barbe, V.; Roche, D.; Cruveiller, S.; et al. The Arginine Deiminase Operon Is Responsible for a Fitness Trade-Off in Extended-Spectrum-β-Lactamase-Producing Strains of Escherichia coli. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Shea, A.E.; Marzoa, J.; Himpsl, S.D.; Smith, S.N.; Zhao, L.; Tran, L.; Mobley, H.L.T. Escherichia coli CFT073 Fitness Factors during Urinary Tract Infection: Identification Using an Ordered Transposon Library. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Roschanski, N.; Hadziabdic, S.; Borowiak, M.; Malorny, B.; Tenhagen, B.A.; Projahn, M.; Kaesbohrer, A.; Guenther, S.; Szabo, I.; Roesler, U.; et al. Detection of VIM-1-Producing Enterobacter cloacae and Salmonella enterica Serovars Infantis and Goldcoast at a Breeding Pig Farm in Germany in 2017 and Their Molecular Relationship to Former VIM-1-Producing S. Infantis Isolates in German Livestock Production. Msphere 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Lv, L.; Huang, X.; Huang, Y.; Zhuang, Z.; Lu, J.; Liu, E.; Wan, M.; Xun, H.; Zhang, Z.; et al. Rapid Increase in Carbapenemase-Producing Enterobacteriaceae in Retail Meat Driven by the Spread of the bla (NDM-5)-Carrying IncX3 Plasmid in China from 2016 to 2018. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Hülter, N.; Sørum, V.; Borch-Pedersen, K.; Liljegren, M.M.; Utnes, A.L.; Primicerio, R.; Harms, K.; Johnsen, P.J. Costs and benefits of natural transformation in Acinetobacter baylyi. BMC Microbiol. 2017, 17, 34. [Google Scholar] [CrossRef] [Green Version]
- Di Luca, M.C.; Sorum, V.; Starikova, I.; Kloos, J.; Hulter, N.; Naseer, U.; Johnsen, P.J.; Samuelsen, O. Low biological cost of carbapenemase-encoding plasmids following transfer from Klebsiella pneumoniae to Escherichia coli. J. Antimicrob. Chemother. 2017, 72, 85–89. [Google Scholar] [CrossRef] [Green Version]
- Cramer, N.; Fischer, S.; Hedtfeld, S.; Dorda, M.; Tümmler, B. Intraclonal competitive fitness of longitudinal cystic fibrosis Pseudomonas aeruginosa airway isolates in liquid cultures. Environ. Microbiol. 2020. [Google Scholar] [CrossRef]
- Smith, M.A.; Bidochka, M.J. Bacterial fitness and plasmid loss: The importance of culture conditions and plasmid size. Can. J. Microbiol. 1998, 44, 351–355. [Google Scholar] [CrossRef]
- Lenski, R.E. Bacterial evolution and the cost of antibiotic resistance. Int. Microbiol. 1998, 1, 265–270. [Google Scholar] [PubMed]
- Dionisio, F.; Conceicao, I.C.; Marques, A.C.; Fernandes, L.; Gordo, I. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 2005, 1, 250–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaufler, K.; Semmler, T.; Pickard, D.J.; de Toro, M.; de la Cruz, F.; Wieler, L.H.; Ewers, C.; Guenther, S. Carriage of Extended-Spectrum Beta-Lactamase-Plasmids Does Not Reduce Fitness but Enhances Virulence in Some Strains of Pandemic E. coli Lineages. Front. Microbiol. 2016, 7, 336. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Scholz, J.; Semmler, T.; Wieler, L.H.; Ewers, C.; Muller, S.; Pickard, D.J.; Schierack, P.; Tedin, K.; Ahmed, N.; et al. ESBL-plasmid carriage in E. coli enhances in vitro bacterial competition fitness and serum resistance in some strains of pandemic sequence types without overall fitness cost. Gut. Pathog. 2018, 10, 24. [Google Scholar] [CrossRef]
- Beceiro, A.; Tomas, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, D.I.; Hughes, D. Effects of Antibiotic Resistance on Bacterial Fitness, Virulence and Transmission. In Evolutionary Biology of Bacterial and Fungal Pathogens; Baquero, F., Nombela, C., Cassell, G.H., Gutiérrez-Fuentes, J.A., Eds.; ASM Press: Washington, WA, USA, 2008; Volume 1, pp. 307–318. [Google Scholar]
- Vogwill, T.; MacLean, R.C. The genetic basis of the fitness costs of antimicrobial resistance: A meta-analysis approach. Evol. Appl. 2015, 8, 284–295. [Google Scholar] [CrossRef]
- Baker, K.S.; Burnett, E.; McGregor, H.; Deheer-Graham, A.; Boinett, C.; Langridge, G.C.; Wailan, A.M.; Cain, A.K.; Thomson, N.R.; Russell, J.E.; et al. The Murray collection of pre-antibiotic era Enterobacteriacae: A unique research resource. Genome Med. 2015, 7, 97. [Google Scholar] [CrossRef] [Green Version]
- Starikova, I.; Al-Haroni, M.; Werner, G.; Roberts, A.P.; Sorum, V.; Nielsen, K.M.; Johnsen, P.J. Fitness costs of various mobile genetic elements in Enterococcus faecium and Enterococcus faecalis. J. Antimicrob. Chemother. 2013, 68, 2755–2765. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Gallego, M.; Torrens, G.; Castillo-Vera, J.; Moya, B.; Zamorano, L.; Cabot, G.; Hultenby, K.; Albertí, S.; Mellroth, P.; Henriques-Normark, B.; et al. Impact of AmpC Derepression on Fitness and Virulence: The Mechanism or the Pathway? Mbio 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Cabot, G.; Bruchmann, S.; Mulet, X.; Zamorano, L.; Moyà, B.; Juan, C.; Haussler, S.; Oliver, A. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob. Agents Chemother. 2014, 58, 3091–3099. [Google Scholar] [CrossRef] [Green Version]
- Williams, M.D.; Ouyang, T.X.; Flickinger, M.C. Starvation-induced expression of SspA and SspB: The effects of a null mutation in sspA on Escherichia coli protein synthesis and survival during growth and prolonged starvation. Mol Microbiol. 1994, 11, 1029–1043. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.M.; Qiu, Y.; Yeh, N.; Blattner, F.R.; Durfee, T.; Jin, D.J. SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol. Microbiol. 2005, 56, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Merrell, D.S.; Camilli, A. Acid tolerance of gastrointestinal pathogens. Curr. Opin. Microbiol. 2002, 5, 51–55. [Google Scholar] [CrossRef]
- Altuvia, S.; Weinstein-Fischer, D.; Zhang, A.; Postow, L.; Storz, G. A small, stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell 1997, 90, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Good, B.H.; McDonald, M.J.; Barrick, J.E.; Lenski, R.E.; Desai, M.M. The dynamics of molecular evolution over 60,000 generations. Nature 2017, 551, 45–50. [Google Scholar] [CrossRef]
- Loftie-Eaton, W.; Yano, H.; Burleigh, S.; Simmons, R.S.; Hughes, J.M.; Rogers, L.M.; Hunter, S.S.; Settles, M.L.; Forney, L.J.; Ponciano, J.M.; et al. Evolutionary Paths That Expand Plasmid Host-Range: Implications for Spread of Antibiotic Resistance. Mol. Biol. Evol. 2016, 33, 885–897. [Google Scholar] [CrossRef] [Green Version]
- De Gelder, L.; Williams, J.J.; Ponciano, J.M.; Sota, M.; Top, E.M. Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics 2008, 178, 2179–2190. [Google Scholar] [CrossRef] [Green Version]
- Heuer, H.; Fox, R.E.; Top, E.M. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiol. Ecol. 2007, 59, 738–748. [Google Scholar] [CrossRef] [Green Version]
- Bouma, J.E.; Lenski, R.E. Evolution of a bacteria/plasmid association. Nature 1988, 335, 351–352. [Google Scholar] [CrossRef]
- San Millan, A.; Toll-Riera, M.; Qi, Q.; MacLean, R.C. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun. 2015, 6, 6845. [Google Scholar] [CrossRef] [Green Version]
- Harrison, E.; Guymer, D.; Spiers, A.J.; Paterson, S.; Brockhurst, M.A. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 2015, 25, 2034–2039. [Google Scholar] [CrossRef] [Green Version]
- Porse, A.; Schonning, K.; Munck, C.; Sommer, M.O. Survival and Evolution of a Large Multidrug Resistance Plasmid in New Clinical Bacterial Hosts. Mol. Biol. Evol. 2016, 33, 2860–2873. [Google Scholar] [CrossRef] [PubMed]
- Clowes, R.C.; Rowley, D. Some observations on linkage effects in genetic recombination in Escherichia coli K-12. J. Gen. Microbiol. 1954, 11, 250–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, B.M.; Harding, G.P.; Zuccarelli, A.J. A general method for detecting and sizing large plasmids. Anal. Biochem. 1995, 226, 235–240. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic. Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Isolate | Wildtype Host Species | Beta-Lactamase Gene | Plasmid Size (kb) | Replicon Type |
|---|---|---|---|---|
| RS039 K1 | E. coli | blaCTX-M-1 | 80 | IncI1 |
| RS292 K1 | E. coli | blaCTX-M-15, blaTEM-1 | 80 | IncFII |
| 17/11 K1 | E. coli | blaKPC-2 | 190 | IncA/C |
| 370/12 K1 | K. oxytoca | blaVIM-1 | 250 | IncFIB |
| 384/13 K2 | K. pneumoniae | blaNDM-1, blaCTX-M-15, blaOXA-9, blaTEM-1 | 110 | IncFII |
| 656/13 K2 | E. coli | blaSHV-12 | 100 | IncI1 |
| 531/12 K2 | E. coli | blaCMY-2 | 90 | IncI1 |
| 252/09 K3 * | E. coli | blaCMY-2 | 85, 60 | IncI1, IncFII |
| 346/12 K2 * | K. pneumoniae | blaOXA-48 | 60 | IncL/M-1 |
| RS165 K1 * | E. coli | blaCTX-M-14 | 70 | IncFII |
| 151/09 K2 * | E. cloacae | blaVIM-1 | 100 | IncN/IncR |
| 104/15 K3 * | E. coli | blaCTX-M-15 | 77 | IncF |
| 102/04 K1 * | E. coli | blaCMY-16, blaTEM-1 | 160 | IncA/C |
| 102/04 K2 * | E. coli | blaCMY-16, blaTEM-1 | 160 | IncA/C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietsch, M.; Pfeifer, Y.; Fuchs, S.; Werner, G. Genome-Based Analyses of Fitness Effects and Compensatory Changes Associated with Acquisition of blaCMY-, blaCTX-M-, and blaOXA-48/VIM-1-Containing Plasmids in Escherichia coli. Antibiotics 2021, 10, 90. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10010090
Pietsch M, Pfeifer Y, Fuchs S, Werner G. Genome-Based Analyses of Fitness Effects and Compensatory Changes Associated with Acquisition of blaCMY-, blaCTX-M-, and blaOXA-48/VIM-1-Containing Plasmids in Escherichia coli. Antibiotics. 2021; 10(1):90. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10010090
Chicago/Turabian StylePietsch, Michael, Yvonne Pfeifer, Stephan Fuchs, and Guido Werner. 2021. "Genome-Based Analyses of Fitness Effects and Compensatory Changes Associated with Acquisition of blaCMY-, blaCTX-M-, and blaOXA-48/VIM-1-Containing Plasmids in Escherichia coli" Antibiotics 10, no. 1: 90. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10010090