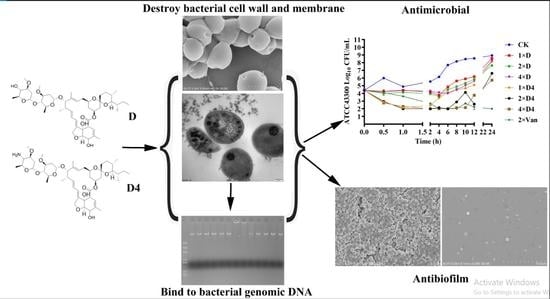
A Novel Ivermectin-Derived Compound D4 and Its Antimicrobial/Biofilm Properties against MRSA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cell Lines
2.2. The Preparation of 4-amino-4-deoxyivermectin B1 (D4)
2.3. Antimicrobial Activity
2.3.1. Minimum Inhibitory Concentration (MIC)
2.3.2. Time-Kill Curves
2.4. Hemolysis and Cytotoxicity
2.4.1. Hemolysis
2.4.2. Cytotoxicity
2.5. Effects of D and D4 on Cell Wall and Membrane
2.5.1. Scanning/Transmission Electron (SEM/TEM) Microscope Observations
2.5.2. Membrane Permeabilization Analysis
2.6. Effects of D and D4 on Bacterial Genomic DNA
2.6.1. Gel Retardation Assay
2.6.2. Circular Dichroism (CD) Spectroscopy
2.7. Ability of D and D4 against MRSA Biofilms
2.7.1. Effects on Biofilm Formation
2.7.2. Biofilms Observed by SEM
2.7.3. Effects on Transcription of Biofilm Formation-Related Genes
2.8. Statistical Analysis
3. Results
3.1. The Characterization of 4-amino-4-deoxyivermectin B1 (D4)
3.2. Antimicrobial Activity
3.2.1. MIC Determination
3.2.2. Time-Killing Curves
3.3. Hemolysis and Cytotoxicity
3.3.1. Hemolysis
3.3.2. Cytotoxicity
3.4. Effects of D and D4 on Cell Wall and Membrane
3.4.1. Scanning/Transmission Electron (SEM/TEM) Microscope Observations
3.4.2. Membrane Permeabilization Analysis
3.5. Effects of D and D4 on Bacterial Genomic DNA
3.5.1. Gel Retardation Assay
3.5.2. CD Spectroscopy
3.6. Ability of D and D4 against MRSA Biofilms
3.6.1. Inhibition of Biofilm Formation
3.6.2. Inhibition of Biofilms Observed by SEM
3.6.3. Effects of D and D4 on the Transcription of Biofilm Formation Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gajdács, M. The Continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics 2019, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Hassoun, A.; Linden, P.K.; Friedman, B. Incidence, prevalence, and management of MRSA bacteremia across patient populations—a review of recent developments in MRSA management and treatment. Crit. Care 2017, 21, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bales, P.M.; Renke, E.M.; May, S.L.; Shen, Y.; Nelson, D.C. Purification and characterization of biofilm-associated EPS exopolysaccharides from ESKAPE organisms and other pathogens. PLoS ONE 2013, 8, 8. [Google Scholar]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, L.; Chan, S.; Hossain, J.; Di Pentima, M.C. Effects of aggregate and individual antibiotic exposure on vancomycin MICs for Staphylococcus aureus isolates recovered from pediatric patients. J. Clin. Microbiol. 2013, 51, 2837–2842. [Google Scholar] [CrossRef] [Green Version]
- Bose, R.J.C.; Tharmalingam, N.; Garcia Marques, F.J.; Sukumar, U.K.; Natarajan, A.; Zeng, Y.; Robinson, E.; Bermudez, A.; Chang, E.; Habte, F.; et al. Reconstructed apoptotic bodies as targeted "Nano Decoys" to treat intracellular bacterial infections within macrophages and cancer cells. ACS Nano. 2020, 14, 5818–5835. [Google Scholar] [CrossRef]
- Boswihi, S.S.; Udo, E.E. Methicillin-resistant Staphylococcus aureus: An update on the epidemiology, treatment options and infection control. Curr. Med. Res. Pr. 2018, 8, 18–24. [Google Scholar] [CrossRef]
- Wilkinson, G.F.; Pritchard, K. In Vitro screening for drug repositioning. J. Biomol. Screen. 2015, 20, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Ashraf, S.; Chaudhry, U.; Raza, A.; Ghosh, D.; Zhao, X. In vitro activity of ivermectin against Staphylococcus aureus clinical isolates. Antimicrob Resist Infect Control 2018, 7, 27. [Google Scholar] [CrossRef]
- Torres, N.S.; Abercrombie, J.J.; Srinivasan, A.; Lopez-Ribot, J.L.; Ramasubramanian, A.K.; Leung, K.P. Screening a commercial library of pharmacologically active small molecules against Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 2016, 60, 5663–5672. [Google Scholar] [CrossRef] [Green Version]
- Gajdács, M.; Spengler, G. The Role of Drug Repurposing in the development of novel antimicrobial drugs: Non-antibiotic pharmacological agents as quorum sensing-inhibitors. Antibiotics 2019, 8, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baraka, O.Z.; Mahmoud, B.M.; Marschk, C.K. Ivermectin distribution in the plasma and tissues of patients infected with Onchocerca volvulus. Eur. J. Clin. Pharmacol. 1996, 50, 407–410. [Google Scholar] [CrossRef]
- Helmut, M.; Philip, E.; Byron, H.A.; Bruce, O.L.; Aino, L.; Alexander, M.; Thomas, L.S.; Maureen, T.; Frank, S.W.; Matthew, J.W.; et al. 4″-Deoxy-4″-aminoavermectins with potent broad spectrum antiparasitic activities. Bioorg. Med. Chem. Lett. 1995, 5, 2435–2440. [Google Scholar]
- Khan, F.I.; Rahman, S.; Queen, A.; Ahamad, S.; Ali, S.; Kim, J.; Hassan, I. Implications of molecular diversity of chitin and its derivatives. Appl. Microbiol. Biotechnol. 2017, 101, 3513–3536. [Google Scholar] [CrossRef]
- Tachaboonyakiat, W.; Sukpaiboon, E.; Pinyakong, O. Development of an antibacterial chitin betainate wound dressing. Polym. J. 2014, 46, 505–510. [Google Scholar] [CrossRef]
- Ren, J.; Wang, P.; Dong, F.; Feng, Y.; Peng, D.; Guo, Z. Synthesis and antifungal properties of 6-amino-6-deoxyinulin, a kind of precursors for facile chemical modifications of inulin. Carbohydr. Polym. 2012, 87, 1744–1748. [Google Scholar] [CrossRef]
- Zhang, J.; Nan, X.; Yu, H.T.; Cheng, P.L.; Zhang, Y.; Liu, Y.Q.; Zhang, S.Y.; Hu, G.F.; Liu, H.X.; Chen, A.L. Synthesis, biological activities and structure-activity relationships for new avermectin analogues. Eur. J. Med. Chem. 2016, 121, 422–432. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Yang, N.; Teng, D.; Mao, R.; Hao, Y.; Wang, X.; Wang, Z.; Wang, X.; Wang, J. A recombinant fungal defensin-like peptide-P2 combats multidrug-resistant Staphylococcus aureus and biofilms. Appl. Microbiol. Biot. 2019, 103, 5193–5213. [Google Scholar] [CrossRef]
- Yang, N.; Li, X.; Teng, D.; Li, Z.; Wang, X.; Mao, R.; Wang, X.; Hao, Y.; Wang, J. Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis. Sci. Rep. 2017, 7, 19. [Google Scholar] [CrossRef]
- An, N.; Cheng, D.H. The long noncoding RNA HOST2 promotes gemcitabine resistance in human pancreatic cancer cells. Pathol. Oncol. Res. 2020, 26, 425–431. [Google Scholar] [CrossRef]
- Wang, X.; Teng, D.; Mao, R.; Yang, N.; Hao, Y.; Wang, J. Combined systems approaches reveal a multistage mode of action of a marine antimicrobial peptide against pathogenic Escherichia coli and its protective effect against bacterial peritonitis and endotoxemia. Antimicrob. Agents Chemother. 2017, 61, 20. [Google Scholar] [CrossRef] [Green Version]
- De Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; de Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; van der Heijde, T.; Boekema, B.K.; et al. Prevention of Staphylococcus aureus biomaterial-associated infections using a polymer-lipid coating containing the antimicrobial peptide OP-145. J. Control. Release 2016, 222, 1–8. [Google Scholar] [CrossRef]
- De Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; De Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; Van Der Heijde, T.; Boekema, B.K.; et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018, 10, eaan4044. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-H.; Park, J.-H.; Cho, H.S.; Joo, S.W.; Cho, M.H.; Lee, J. Anti-biofilm activities of quercetin and tannic acid against Staphylococcus aureus. Biofouling 2013, 29, 491–499. [Google Scholar] [CrossRef]
- Chung, P.Y. Novel targets of pentacyclic triterpenoids in Staphylococcus aureus: A systematic review. Phytomedicine 2020, 73, 152933. [Google Scholar] [CrossRef]
- Archer, N.K.; Mazaitis, M.J.; Costerton, J.W.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms Properties, regulation and roles in human disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [Green Version]
- Garrido, V.; Collantes, M.; Barberán, M.; Peñuelas, I.; Arbizu, J.; Amorena, B.; Grilló, M.J. In vivo monitoring of Staphylococcus aureus biofilm infections and antimicrobial therapy by [18F]fluoro-deoxyglucose-MicroPET in a mouse model. Antimicrob Agents Chemother. 2014, 58, 6660–6667. [Google Scholar] [CrossRef] [Green Version]
- Brackman, G.; De Meyer, L.; Nelis, H.; Coenye, T. Biofilm inhibitory and eradicating activity of wound care products against Staphylococcus aureus and Staphylococcus epidermidis biofilms in an in vitro chronic wound model. J. Appl. Microbiol. 2013, 114, 1833–1842. [Google Scholar] [CrossRef]
- Gajdács, M.; Ábrók, M.; Lázár, A.; Burián, K. Increasing relevance of Gram-positive cocci in urinary tract infections: A 10-year analysis of their prevalence and resistance trends. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Arias, M.; Piga, K.B.; Hyndman, M.E.; Vogel, H.J. Improving the activity of Trp-rich antimicrobial peptides by Arg/Lys substitutions and changing the length of cationic residues. Biomology 2018, 8, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaoru, Y.; Yukio, A.; Masaki, S.; Mitsuko, O.; Katsushiro, M.; Takahiro, T.; Yukihiro, A.; Kuniko, T.; Ka-zunori, T. Direct anti-biofilm effects of macrolides on Acinetobacter baumannii: Comprehensive and comparative demonstration by a simple assay using microtiter plate combined with peglid. Biomed. Res. 2020, 41, 259–268. [Google Scholar]
- Del Pozo, J.L. Biofilm-related disease. Expert Rev. Anti-infective Ther. 2018, 16, 51–65. [Google Scholar] [CrossRef]
- Hobbs, J.K.; Boraston, A.B. (p)ppGpp and the stringent response: An emerging threat to antibiotic therapy. ACS Infect. Dis. 2019, 5, 1505–1517. [Google Scholar] [CrossRef]
- Lauderdale, K.J.; Boles, B.R.; Cheung, A.L.; Horswill, A.R. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 2009, 77, 1623–1635. [Google Scholar] [CrossRef] [Green Version]
- Goudarzi, M.; Mohammadi, A.; Amirpour, A.; Fazeli, M.; Nasiri, M.J.; Hashemi, A.; Goudarzi, H. Genetic diversity and biofilm formation analysis of Staphylococcus aureus causing urinary tract infections in Tehran, Iran. J. Infect. Dev. Ctries. 2019, 13, 777–785. [Google Scholar] [CrossRef] [Green Version]
- Omidi, M.; Firoozeh, F.; Saffari, M.; Sedaghat, H.; Zibaei, M.; Khaledi, A. Ability of biofilm production and molecular analysis of spa and ica genes among clinical isolates of methicillin-resistant Staphylococcus aureus. BMC Res. Notes 2020, 13, 1–7. [Google Scholar] [CrossRef] [Green Version]





| Gene | Sequence (5′ to 3′) |
|---|---|
| RSH-F | TACATCGCACTGATTGCCCA |
| RSH-R | TTAAATTGCCGGCTGTCGAG |
| relP-F | TTGCCGGAATTCGCGTAGTA |
| relP-R | CGCGTTCTGCTAAAAAGACTGG |
| relQ-F | AGAAAGTGGTTACCGCTCGT |
| relQ-R | TCATCCGGATAAGCACCATCA |
| rsbU-F | CGCGTGAAGATGTGTTCAAGAC |
| rsbU-R | CTATCTCTTTATCGTGAACTTGAAG |
| sigB-F | GGTGCCATAAATAGATTCGATATGTCCTT |
| sigB-R | CTTTTGATTTCACCGATTACAGTAGGTACT |
| spA-F | GCGCAACACGATGAAGCTCAACAA |
| spA-R | ACGTTAGCACTTTGGCTTGGATCA |
| AgrA-F | AAGCATGACCCAGTTGGTAACA |
| AgrA-R | ATCCATCGCTGCAACTTTGTAGA |
| icaD-F | ATGGTCAAGCCCAGACAGAG |
| icaD-R | AGTATTTTCAATGTTTAAAGCAA |
| 16s rRNA-F | GCTGCCCTTTGTATTGTC |
| 16s rRNA-R | AGATGTTGGGTTAAGTCCC |
| Drugs | MIC (μg/mL) |
|---|---|
| ATCC 43300 | |
| D | 20 |
| D4 | 5 |
| vancomycin | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, X.; Xie, H.; Zhang, B.; Zhou, J.; Dou, Z.; Wang, X.; Wang, N. A Novel Ivermectin-Derived Compound D4 and Its Antimicrobial/Biofilm Properties against MRSA. Antibiotics 2021, 10, 208. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10020208
Tan X, Xie H, Zhang B, Zhou J, Dou Z, Wang X, Wang N. A Novel Ivermectin-Derived Compound D4 and Its Antimicrobial/Biofilm Properties against MRSA. Antibiotics. 2021; 10(2):208. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10020208
Chicago/Turabian StyleTan, Xinyi, Haoji Xie, Bin Zhang, Jiale Zhou, Zhende Dou, Xiao Wang, and Ning Wang. 2021. "A Novel Ivermectin-Derived Compound D4 and Its Antimicrobial/Biofilm Properties against MRSA" Antibiotics 10, no. 2: 208. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10020208