Characterization of Extended-Spectrum β-Lactamase-Producing and AmpC β-Lactamase-Producing Enterobacterales Isolated from Companion Animals in Korea
Abstract
:1. Introduction
2. Results
2.1. ESC Resistance Gene Detection
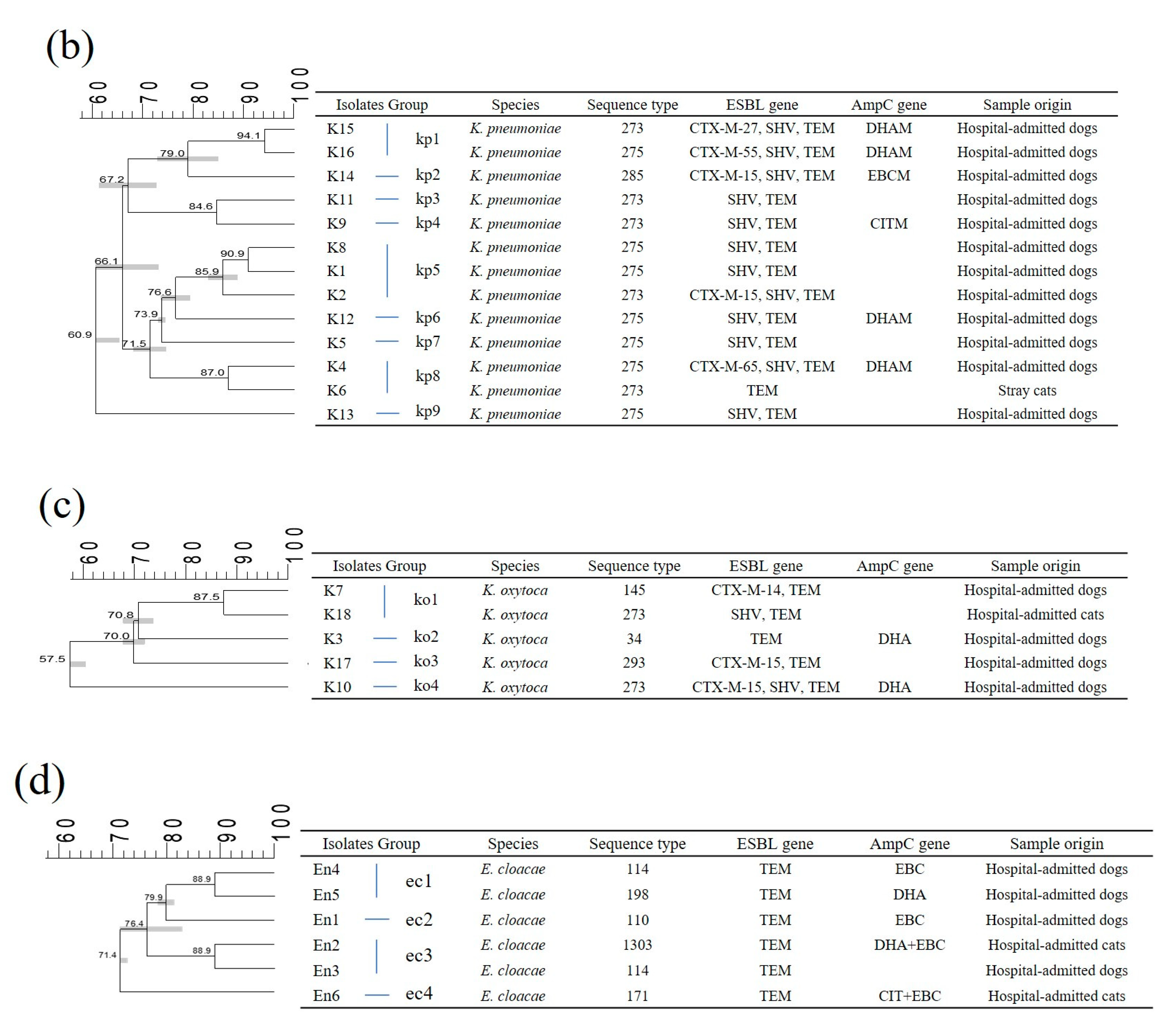
2.2. Multi-Locus Sequence Typing (MLST)
2.3. Pulsed-Field Gel Electrophoresis (PFGE)
2.4. Genetic Relatedness
3. Discussion
4. Materials and Methods
4.1. Bacterial Characterization
4.2. Characterization of β-Lactamase Genes
4.3. Multi-Locus Sequence Typing
4.4. Pulsed-Field Gel Electrophoresis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ur Rahman, S.; Ali, T.; Ali, I.; Khan, N.A.; Han, B.; Gao, J. The growing genetic and functional diversity of extended spectrum β-lactamases. BioMed Res. Int. 2018, 2018, 9519718. [Google Scholar] [CrossRef]
- Sheng, W.-H.; Badal, R.E.; Hsueh, P.-R.; Program, S. Distribution of extended-spectrum β-lactamases, AmpC β-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: Results of the study for monitoring antimicrobial resistance trends (SMART). Antimicrob. Agents Chemother. 2013, 57, 2981–2988. [Google Scholar] [CrossRef] [Green Version]
- Sougakoff, W.; Goussard, S.; Gerbaud, G.; Courvalin, P. Plasmid-Mediated Resistance to Third-Generation Cephalosporins Caused by Point Mutations in TEM-Type Penicillinase Genes. Rev. Infect. Dis. 1988, 10, 879–884. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Ikeda, F.; Kamimura, T.; Yokota, Y.; Mine, Y. Novel plasmid-mediated beta-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob. Agents Chemother. 1988, 32, 1243–1246. [Google Scholar] [CrossRef] [Green Version]
- Rossolini, G.M.; D’Andrea, M.M.; Mugnaioli, C. The spread of CTX-M-type extended-spectrum β-lactamases. Clin. Microbiol. Infect. Off. Publ. Europ. Soc. Clin. Microbiol. Infect. Dis. 2008, 14 (Suppl. 1), 33–41. [Google Scholar] [CrossRef] [Green Version]
- Tamang, M.D.; Nam, H.-M.; Jang, G.-C.; Kim, S.-R.; Chae, M.H.; Jung, S.-C.; Byun, J.-W.; Park, Y.H.; Lim, S.-K. Molecular characterization of extended-spectrum-β-lactamase-producing and plasmid-mediated AmpC β-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob. Agents Chemother. 2012, 56, 2705–2712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Lee, H.; Lee, K.; Jeong, S.H.; Bae, I.K.; Kim, J.S.; Kwak, H.S. CTX-M-14 and CTX-M-15 enzymes are the dominant type of extended-spectrum β-lactamase in clinical isolates of Escherichia coli from Korea. J. Med. Microbiol. 2009, 58, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Philippon, A.; Arlet, G.; Jacoby, G.A. Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 2002, 46, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.S.; Song, W.; Park, H.-M.; Oh, J.-Y.; Chae, J.-C.; Shin, S.; Jeong, S.H. Clonal spread of extended-spectrum cephalosporin-resistant Enterobacteriaceae between companion animals and humans in South Korea. Front. Microbiol. 2019, 10, 1371. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.g.; Jeong, J.; Kim, M.j.; Park, D.w.; Shin, J.h.; Park, H.j.; Chung, J.k.; Kee, H.y. Prevalence and molecular epidemiology of ESBLs, plasmid-determined AmpC-type β-lactamases and carbapenemases among diarrhoeagenic Escherichia coli isolates from children in Gwangju, Korea: 2007–16. J. Antimicrob. Chemother. 2019, 74, 2181–2187. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-J.; Moon, D.C.; Mechesso, A.F.; Kang, H.Y.; Kim, M.H.; Choi, J.-H.; Kim, S.-J.; Yoon, S.-S.; Lim, S.-K. Resistance profiling and molecular characterization of extended-spectrum/plasmid-mediated AmpC β-lactamase-producing Escherichia coli isolated from healthy broiler chickens in South Korea. Microorganisms 2020, 8, 1434. [Google Scholar] [CrossRef]
- Shin, S.W.; Jung, M.; Shin, M.-K.; Yoo, H.S. Profiling of antimicrobial resistance and plasmid replicon types in β-lactamase producing Escherichia coli isolated from Korean beef cattle. J. Vet. Sci. 2015, 16, 483–489. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial resistance: A one health perspective. Antimicrob. Resist. Bact. Livest. Compan. Anim. 2018, 6, 521–547. [Google Scholar]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. Off. Publ. Europ. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 646–655. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Choi, M.-J.; Ko, K.S. Replicon sequence typing of IncF plasmids and the genetic environments of blaCTX-M-15 indicate multiple acquisitions of blaCTX-M-15 in Escherichia coli and Klebsiella pneumoniae isolates from South Korea. J. Antimicrob. Chemother. 2012, 67, 1853–1857. [Google Scholar] [CrossRef] [Green Version]
- Coque, T.M.; Novais, A.; Carattoli, A.; Poirel, L.; Pitout, J.; Peixe, L.; Baquero, F.; Cantón, R.; Nordmann, P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 2008, 14, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.A.; Park, Y.S.; Kim, H.; Seo, Y.H.; Lee, K. The usefulness of active surveillance culture of extended-spectrum β-lactamase-producing Escherichia coli in ICU settings without outbreak in the situation of wide spread of sequence type 131 ESBL-producing E. coli in community. Ann. Clin. Microbiol. Vol. 2018, 21, 1096937. [Google Scholar] [CrossRef]
- Donati, V.; Feltrin, F.; Hendriksen, R.S.; Svendsen, C.A.; Cordaro, G.; García-Fernández, A.; Lorenzetti, S.; Lorenzetti, R.; Battisti, A.; Franco, A. Extended-spectrum-β-lactamases, AmpC β-lactamases and plasmid mediated quinolone resistance in klebsiella spp. from companion animals in Italy. PLoS ONE 2014, 9, e90564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mammina, C.; Bonura, C.; Di Bernardo, F.; Aleo, A.; Fasciana, T.; Sodano, C.; Saporito, M.; Verde, M.; Tetamo, R.; Palma, D. Ongoing spread of colistin-resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Eurosurveillance 2012, 17, 20248. [Google Scholar]
- Silva, Y.; Gomes Ferrari, R.; Marin, V.; Conte Junior, C. A global overview of β-lactam resistance genes in Klebsiella pneumoniae. Open Infect. Dis. J. 2019, 11, 22–34. [Google Scholar] [CrossRef]
- Kao, C.-Y.; Udval, U.; Huang, Y.-T.; Wu, H.-M.; Huang, A.-H.; Bolormaa, E.; Yan, J.-J.; Urangoo, Z.; Batbaatar, G.; Khosbayar, T. Molecular characterization of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella spp. isolates in Mongolia. J. Microbiol. Immunol. Infect. 2016, 49, 692–700. [Google Scholar] [CrossRef] [Green Version]
- Schmiedel, J.; Falgenhauer, L.; Domann, E.; Bauerfeind, R.; Prenger-Berninghoff, E.; Imirzalioglu, C.; Chakraborty, T. Multiresistant extended-spectrum β-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiol. 2014, 14, 187. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.H.; Lee, J.E.; Park, S.J.; Kim, M.N.; Choo, E.J.; Kwak, Y.G.; Jeong, J.Y.; Woo, J.H.; Kim, N.J.; Kim, Y.S. Prevalence, microbiology, and clinical characteristics of extended-spectrum β-lactamase-producing Enterobacter spp., Serratia marcescens, Citrobacter freundii, and Morganella morganii in Korea. Europ. J. Clin. Microb. Infect. Dis. 2007, 26, 557–561. [Google Scholar] [CrossRef]
- Jung, W.K.; Shin, S.; Park, Y.K.; Lim, S.-K.; Moon, D.-C.; Park, K.T.; Park, Y.H. Distribution and antimicrobial resistance profiles of bacterial species in stray cats, hospital-admitted cats, and veterinary staff in South Korea. BMC Vet. Res. 2020, 16, 109. [Google Scholar] [CrossRef] [Green Version]
- Jung, W.K.; Shin, S.; Park, Y.K.; Noh, S.M.; Shin, S.R.; Yoo, H.S.; Park, S.C.; Park, Y.H.; Park, K.T. Distribution and antimicrobial resistance profiles of bacterial species in stray dogs, hospital-admitted dogs, and veterinary staff in South Korea. Prev. Vet. Med. 2020, 184, 105151. [Google Scholar] [CrossRef]
- Lee, H.; Yoon, E.-J.; Kim, D.; Jeong, S.H.; Shin, J.H.; Shin, J.H.; Shin, K.S.; Kim, Y.A.; Uh, Y.; Park, C.; et al. Establishment of the South Korean national antimicrobial resistance surveillance system, Kor-GLASS, in 2016. Eurosurveillance 2018, 23, 1700734. [Google Scholar] [CrossRef] [PubMed]
- Thabit, A.G.; El-Khamissy, T.R.; Ibrahim, M.A.; Attia, A.E. Detection of extended-spectrum β-lactamase enzymes (ESBLs) produced by Escherichia coli urinary pathogens at assiut university hospital. Bullet. Pharm. Sci. 2011, 34, 93–103. [Google Scholar]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyoshi-Akiyama, T.; Hayakawa, K.; Ohmagari, N.; Shimojima, M.; Kirikae, T. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS ONE 2013, 8, e66358. [Google Scholar] [CrossRef] [Green Version]
- Tartof, S.Y.; Solberg, O.D.; Manges, A.R.; Riley, L.W. Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J. Clin. Microbiol. 2005, 43, 5860–5864. [Google Scholar] [CrossRef] [Green Version]
- Diancourt, L.; Passet, V.; Verhoef, J.; Grimont, P.A.; Brisse, S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005, 43, 4178–4182. [Google Scholar] [CrossRef] [Green Version]
- Ribot, E.M.; Fair, M.A.; Gautom, R.; Cameron, D.N.; Hunter, S.B.; Swaminathan, B.; Barrett, T.J. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for pulsenet. Foodborne Pathog. Dis. 2006, 3, 59–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Organism | Hospital-Admitted Dogs (n = 56) | Stray Dogs (n = 23) | Hospital-Admitted Cats (n = 11) | Stray Cats (n = 1) | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaCTX-M | blaSHV | blaTEM | blaCTX-M | blaSHV | blaTEM | blaCTX-M | blaSHV | blaTEM | blaCTX-M | blaSHV | blaTEM | blaCTX-M | blaSHV | blaTEM | |
| E. coli (n = 51) | 21 | − | 22 | 15 | − | 23 | 6 | − | 6 | − | − | − | 42 (82.4%) | − | 51 (100%) |
| K. pneumoniae (n = 17) | 7 | 15 | 15 | − | − | − | 1 | 1 | 1 | 1 | − | 1 | 9 (17.6%) | 16 (94.1%) | 17 (100%) |
| K. oxytoca (n = 5) | 4 | 1 | 4 | − | − | − | − | 1 | 1 | − | − | − | 4 (80.0%) | 2 (40.0%) | 5 (100%) |
| S. marcescens (n = 4) | − | 1 | 3 | − | − | − | − | − | 1 | − | − | − | − | 1 (45.0%) | 4 (100%) |
| S. liquefaciens (n = 7) | − | − | 7 | − | − | − | − | − | − | − | − | − | − | − | 7 (100%) |
| E. cloacae (n = 7) | − | − | 5 | − | − | − | − | − | 2 | − | − | − | − | − | 7 (100%) |
| Organism | Sample Origin | blaCTX-M Subtype Group | Unidentified | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX-M-1 | CTX-M-9 | ||||||||||
| CTX-M-15 | CTX-M-55 | CTX-M-3 | CTX-M-61 | Total | CTX-M-14 | CTX-M-27 | CTX-M-65 | Total | |||
| E. coli (n = 42) | Hospital-admitted dogs (n = 21) | 7 | 6 | 1 | 1 | 15 | 3 | 1 | − | 4 | 2 |
| Stray dogs (n = 15) | 8 | 3 | − | − | 11 | 3 | 1 | − | 4 | 0 | |
| Hospital-admitted cats (n = 6) | 2 | 2 | − | − | 4 | − | − | − | 0 | 2 | |
| K. pneumoniae (n = 9) | Hospital-admitted dogs (n = 7) | 4 | 1 | − | − | 5 | − | 1 | 1 | 2 | 0 |
| Hospital-admitted cats (n = 1) | − | − | 1 | − | 1 | − | − | − | 0 | 0 | |
| Stray cats (n = 1) | − | − | − | − | 0 | − | − | − | 0 | 1 | |
| K. oxytoca (n = 4) | Hospital-admitted dogs (n = 4) | 2 | − | − | − | 2 | 1 | − | − | 1 | 1 |
| Total | 23 (41.8%) | 12 (21.8%) | 2 (3.6%) | 1 (1.8%) | 38 (69.1%) | 7 (12.7%) | 3 (5.5%) | 1 (1.8%) | 11 (20.0%) | 6 (10.9%) | |
| Organism | Sample Origin | AmpC β-Lactamases Gene | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MOX | CIT | DHA | ACC | EBC | FOX | EBC+CIT | EBC+DHA | Total | ||
| E. coli | Hospital-admitted dogs (n = 22) | − | 5 | − | − | − | − | − | − | 5 |
| Stray dogs (n = 23) | − | 13 | − | − | − | 1 | − | − | 14 | |
| K. pneumoniae | Hospital-admitted dogs (n = 15) | − | 1 | 4 | − | 1 | − | − | − | 6 |
| Hospital-admitted cats (n = 1) | − | − | 1 | − | − | − | − | − | 1 | |
| K. oxytoca | Hospital-admitted dogs (n = 4) | − | − | 2 | − | − | − | − | − | 2 |
| E. cloacae | Hospital-admitted dogs (n = 5) | − | − | 1 | − | 2 | − | − | − | 3 |
| Hospital-admitted cats (n = 2) | − | − | − | − | − | − | 1 | 1 | 2 | |
| Total | − | 19 | 8 | − | 3 | 1 | 1 | 1 | 33 | |
| Organism | blaCTX-M Cluster | ESC Resistance Gene | No. Isolation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ESBLs | AmpCs | ST Type | |||||||
| blaCTX-M | blaSHV | blaTEM | Hospital-Admitted Dogs | Stray Dogs | Hospital-Admitted Cats | Stray Cats | |||
| E. coli | CTX-M-1 | CTX-M-15 | − | + | − | 648 (n = 1) | 354 (n = 1) | 131 (n = 1) | − |
| 1981 (n = 1) | 448 (n = 2) | 1262 (n = 1) | − | ||||||
| 2179 (n = 1) | 7644 (n = 1) | − | − | ||||||
| 10,207 (n = 1) | 10,459 (n = 1) | − | − | ||||||
| − | + | CIT | 457 (n = 1) | 354 (n = 2) | − | − | |||
| 5667 (n = 1) | 410 (n = 1) | − | − | ||||||
| 11,000 (n = 1) | − | − | − | ||||||
| CTX-M-55 | − | + | − | 4616 (n = 1) | − | 131 (n = 1) | − | ||
| 5150 (n = 1) | − | 156 (n = 1) | − | ||||||
| 8451 (n = 1) | − | − | − | ||||||
| 8908 (n = 1) | − | − | − | ||||||
| − | + | CIT | 2505 (n = 1) | 410 (n = 1) | − | − | |||
| 3285 (n = 1) | 3285 (n = 2) | − | − | ||||||
| CTX-M-9 | CTX-M-61 | − | + | − | 3285 (n = 1) | − | − | − | |
| CTX-M-3 | − | + | − | 8885 (n = 1) | − | − | − | ||
| CTX-M-14 | − | + | − | 10,220 (n = 1) | 38 (n = 1) | − | − | ||
| 131 (n = 2) | 68 (n = 1) | − | − | ||||||
| − | 648 (n = 1) | − | − | ||||||
| CTX-M-27 | − | + | − | 131 (n = 1) | − | − | − | ||
| CTX-M-27 | − | + | FOX | − | 1193 (n = 1) | − | − | ||
| − | − | − | − | ||||||
| Unidentified | − | − | + | − | 131 (n = 2) | − | 156 (n = 1) | − | |
| − | − | 6105 (n = 1) | − | ||||||
| Negative | − | − | + | − | 372 (n = 1) | 2541 (n = 1) | − | − | |
| − | − | + | CIT | − | 405 (n = 5) | − | − | ||
| − | 457 (n = 2) | − | − | ||||||
| Total | 22 | 23 | 6 | 0 | |||||
| K. pneumoniae | CTX-M-1 | CTX-M-15 | + | + | − | 273 (n = 2) | − | − | − |
| 275 (n = 1) | − | − | − | ||||||
| + | + | EBC | 285 (n = 1) | − | − | − | |||
| CTX-M-55 | + | + | DHA | 275 (n = 1) | − | − | − | ||
| CTX-M-3 | + | + | DHA | − | − | 273 (n = 1) | − | ||
| CTX-M-9 | CTX-M-27 | + | + | DHA | 273 (n = 1) | − | − | − | |
| CTX-M-65 | + | + | DHA | 275 (n = 1) | − | − | − | ||
| Unidentified | − | - | + | − | − | − | 273 (n = 1) | ||
| Negative | − | + | + | − | 273 (n = 2) | − | − | − | |
| 275 (n = 4) | − | − | − | ||||||
| − | + | + | CIT | 273 (n = 1) | − | − | − | ||
| − | + | + | DHA | 275 (n = 1) | − | − | − | ||
| Total | 15 | 0 | 1 | 1 | |||||
| K. oxytoca | CTX-M-1 | CTX-M-15 | − | + | − | 293 (n = 1) | − | − | − |
| + | + | DHA | 273 (n = 1) | − | − | − | |||
| CTX-M-9 | CTX-M-14 | − | + | − | 145 (n = 1) | − | − | − | |
| Unidentified | − | − | + | DHA | 34 (n = 1) | − | − | − | |
| Negative | − | + | + | − | − | − | 273 (n = 1) | − | |
| Total | 4 | 0 | 1 | 0 | |||||
| S. liquefaciens | Negative | − | − | + | − | a ND (n = 7) | − | − | − |
| S. marcescens | Negative | − | − | + | − | ND (n = 2) | − | ND (n = 1) | − |
| + | + | − | ND (n = 1) | − | − | − | |||
| Total | 10 | 0 | 1 | 0 | |||||
| E. cloacae | Negative | − | − | + | DHA | 198 (n = 1) | − | − | − |
| − | − | + | EBC | 114 (n = 1) | − | − | − | ||
| 110 (n = 1) | − | − | − | ||||||
| − | − | + | CIT+EBC | − | − | 171 (n = 1) | − | ||
| − | − | + | DHA+EBC | − | − | 1303 (n = 1) | − | ||
| − | − | + | − | 1252 (n = 1) | − | − | − | ||
| 114 (n = 1) | − | − | − | ||||||
| Total | 5 | 0 | 2 | 0 | |||||
| Organism | Origin | Antimicrobial Resistant Rate against Cephalosporins (%) | Reference | ||||
|---|---|---|---|---|---|---|---|
| Cephalexin | Cefoxitin | Ceftiofur | Ceftriaxone | Cephalothin | |||
| E. coli (n = 51) | Hospital-admitted dogs (n = 22) | 95.5 | 27.3 | 95.5 | 95.5 | 100 | [25] |
| Stray dogs (n = 23) | 100 | 47.8 | 100 | 100 | 100 | ||
| Hospital-admitted cats (n = 6) | 100 | 16.7 | 100 | 100 | 100 | [24] | |
| K. pneumonia (n = 17) | Hospital-admitted dogs (n = 15) | 53.3 | 60.0 | 33.3 | 40 | 53.3 | [25] |
| Hospital-admitted cats (n = 1) | 100 | 100 | 100 | 100 | 100 | [24] | |
| Stray cat (n = 1) | 100 | 100 | 100 | 100 | 100 | ||
| K. oxytoca (n = 5) | Hospital-admitted dogs (n = 4) | 75.0 | 50.0 | 100 | 100 | 100 | [25] |
| Hospital-admitted cat (n = 1) | 100 | 100 | 0 | 0 | 100 | [24] | |
| S. marcescens (n = 4) | Hospital-admitted dogs (n = 3) | 100 | 100 | 0 | 0 | 100 | [25] |
| Hospital-admitted cat (n = 1) | 100 | 100 | 0 | 0 | 100 | [24] | |
| S. liquefaciens (n = 7) | Hospital-admitted dogs (n = 7) | 0 | 14.3 | 28.6 | 0 | 100 | [25] |
| E. cloacae (n = 7) | Hospital-admitted dogs (n = 5) | 100 | 100 | 60.0 | 80.0 | 100 | [25] |
| Hospital-admitted cats (n = 2) | 100 | 100 | 100 | 100 | 100 | [24] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, S.R.; Noh, S.M.; Jung, W.K.; Shin, S.; Park, Y.K.; Moon, D.C.; Lim, S.-K.; Park, Y.H.; Park, K.T. Characterization of Extended-Spectrum β-Lactamase-Producing and AmpC β-Lactamase-Producing Enterobacterales Isolated from Companion Animals in Korea. Antibiotics 2021, 10, 249. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10030249
Shin SR, Noh SM, Jung WK, Shin S, Park YK, Moon DC, Lim S-K, Park YH, Park KT. Characterization of Extended-Spectrum β-Lactamase-Producing and AmpC β-Lactamase-Producing Enterobacterales Isolated from Companion Animals in Korea. Antibiotics. 2021; 10(3):249. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10030249
Chicago/Turabian StyleShin, Se Ra, Seong Mi Noh, Woo Kyung Jung, Sook Shin, Young Kyung Park, Dong Chan Moon, Suk-Kyung Lim, Yong Ho Park, and Kun Taek Park. 2021. "Characterization of Extended-Spectrum β-Lactamase-Producing and AmpC β-Lactamase-Producing Enterobacterales Isolated from Companion Animals in Korea" Antibiotics 10, no. 3: 249. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10030249






