Antagonistic Activity of Bacteria Isolated from the Periplaneta americana L. Gut against Some Multidrug-Resistant Human Pathogens
Abstract
:1. Introduction
2. Materials and Methods
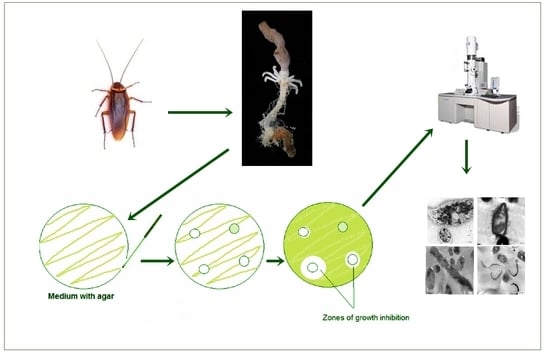
2.1. Collection of Cockroaches and Extraction of Gut Bacteria
2.1.1. Sample Collection
2.1.2. Isolation and Identification of the Gut-Associated Bacteria
2.2. Antagonistic Activity of the Gut Associated Bacteria
2.3. Transmission Electron Microscopy (TEM)
3. Results
3.1. Identification of Isolated Gut Bacteria by Using Vitek (MALDI-TOF MS)
3.2. Evaluating the Antagonistic Activity of Isolated Gut Bacteria
3.3. Ultrastructural Changes Shown by the Tested Pathogens Due to the Antagonistic Effects of Bacterial Symbionts
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [Green Version]
- Odonkor, S.T.; Addo, K.K. Bacteria resistance to antibiotics: Recent trends and challenges. Int. J. Biol. Med. Res. 2011, 2, 1204–1210. [Google Scholar]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st Century. Perspect. Med. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Siddiqui, R.; Khan, N.A. Animals living in polluted environments are potential source of antimicrobials against infec-tious agents. Pathog. Glob. Health 2012, 106, 218–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chellaram, C.; Praveen, M.M. Molecular characterization of antagonistic bacteria, Pseudomonas otitidis from Insect gut, short horned grasshopper. J. Pure Appl. Microbiol. 2015, 9, 2391–2396. [Google Scholar]
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Dillon, R.J.; Dillon, V.M. The gut bacteria of insects: Nonpathogenic interactions. Ann. Rev. Ent. 2004, 49, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Voirol, L.R.P.; Frago, E.; Kaltenpoth, M.; Hilker, M.; Fatouros, N.E. Bacterial symbionts in lepidoptera: Their Diversity, transmission, and impact on the host. Front. Microbiol. 2018, 9, 556. [Google Scholar] [CrossRef] [PubMed]
- Beccaloni, G.W. Cockroach Species File Online, Version 5.0/5.0. World Wide Web Electronic Publication. 2014. Available online: http://Cockroach.SpeciesFile.org (accessed on 20 July 2020).
- Akbar, N.; Siddiqui, R.; Iqbal, M.; Sagathevan, K.; Khan, N.A. Gut bacteria of cockroaches are a potential source of antibacterial compound(s). Lett. Appl. Microbiol. 2018, 66, 416–426. [Google Scholar] [CrossRef]
- Tinker, K.A.; Ottesen, E.A. The core gut microbiome of the american cockroach, Periplaneta americana, is stable and resilient to dietary shifts. Appl. Environ. Microbiol. 2016, 82, 6603–6610. [Google Scholar] [CrossRef] [Green Version]
- Cruden, D.L.; Markovetz, A.J. Microbial aspects of the cockroach hindgut. Arch. Microbiol. 1984, 138, 131–139. [Google Scholar] [CrossRef]
- Gijzen, H.J.; Barugahare, M. Contribution of anaerobic protozoa and methanogens to hindgut metabolic activities of the American cockroach, Periplaneta americana. Appl. Environ. Microbiol. 1992, 58, 2565–2570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzman, J.; Vilcinskas, A. Bacteria associated with cockroaches: Health risk or biotechnological opportunity? Appl. Microbiol. Biotechnol. 2020, 104, 10369–10387. [Google Scholar] [CrossRef]
- Rust, M.K.; Reierson, D.A.; Hansgen, K.H. Control of american cockroaches (Dictyoptera: Blattidae) in sewers. J. Med. Èntomol. 1991, 28, 210–213. [Google Scholar] [CrossRef]
- William, J.; Adiyodi, K.G. American Cockroach; Springer: Berlin/Heidelberg, Germany, 1981; pp. 1–4. [Google Scholar]
- Schapheer, C.; Lopez-Uribe, M.M.; Vera, A.; Villagra, C.A. Distribution, habitat use and plant associations of Moluchia brevipennis (Saussure, 1864) (Blattodea: Ectobiidae): An endemic cockroach from Chilean Mediterranean Matorral biome. Rev. Bras. Èntomol. 2017, 61, 114–122. [Google Scholar] [CrossRef]
- Cochran, D.G. Blattodea:(Cockroaches). In Encyclopedia of Insects; Ring, C., Resh, V.H., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 108–112. [Google Scholar]
- Dietrich, C.; Köhler, T.; Brune, A. The cockroach origin of the termite gut microbiota: Patterns in bacterial community structure reflect major evolutionary events. Appl. Environ. Microbiol. 2014, 80, 2261–2269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kesselheim, A.; Outterson, K.S. Fighting antibiotic resistance: Marrying new financial incentives to meeting public health goals. Health Aff. 2010, 29, 1689–1696.–1696. [Google Scholar] [CrossRef] [Green Version]
- Cascio, A.; Mezzatesta, M.L.; Odierna, A.; Di Bernardo, F.; Barberi, G.; Iaria, C.; Stefani, S.; Giordano, S. Extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacter cloacae ventriculitis successfully treated with intra-ventricular colistin. Int. J. Infect. Dis. 2014, 20, 66–67. [Google Scholar] [CrossRef] [Green Version]
- Dijk, Y.; Bik, E.; Hochstenbach-Vernooij, S.; Vlist, G.; Savelkoul, P.; Kaan, J.; Diepersloot, R. Management of an outbreak of Enterobacter cloacae in a neonatal unit using simple preventive measures. J. Hosp. Infect. 2002, 51, 21–26. [Google Scholar] [CrossRef]
- Daigle, F. Typhi genes expressed during infection or involved in pathogenesis. J. Infect. Dev. Ctries. 2008, 2, 431–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, R.J.; Morris-Jones, R. Bacterial Infections. Rook’s Textbook of Dermatology, 9th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2016; pp. 1–100. [Google Scholar]
- Krzyściak, W.; Jurczak, A.; Kościelniak, D.; Bystrowska, B.; Skalniak, A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 499–515. [Google Scholar] [CrossRef] [Green Version]
- Zafar, N.; Ali, A.; Afzal, M.Y.; Tanveer, Q.; Bibi, S.; Basit, I.; Nasir, H.; Imtiaz, S.; Nazir, U. Role of Streptococci as etiological agents of dental caries. Nov. Res. Microbiol. J. 2020, 4, 766–778. [Google Scholar] [CrossRef]
- Hedayati, M.T.; Pasqualotto, A.C.; Warn, P.A.; Bowyer, P.; Denning, D.W. Aspergillus flavus: Human pathogen, allergen and mycotoxin producer. Microbiology 2007, 153, 1677–1692. [Google Scholar] [CrossRef] [Green Version]
- Walsh, T.J.; Anaissie, E.J.; Denning, D.W.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Patterson, T.F. Treatment of asper-gillosis: Clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 2008, 46, 327–360. [Google Scholar] [CrossRef] [PubMed]
- Abarca, M.L.; Bragulat, M.R.; Castellá, G.; Cabañes, F.J. Ochratoxin A production by strains of Aspergillus niger var. niger. Appl. Environ. Microbiol. 1994, 60, 2650–2652. [Google Scholar] [CrossRef] [Green Version]
- Dutta, A. Candidiasis. In Introduction to Clinical Infectious Diseases; Domachowske, J., Ed.; Springer International Publishing: Geneva, Switzerland, 2019; pp. 335–340. [Google Scholar]
- Moustafa, S. Possibility of reducing presence of harmful fungi in air-conditioner windows using a transcendental anti-fungal chemical. Egypt. Acad. J. Biol. Sci. G. Microbiol. 2019, 11, 59–70. [Google Scholar]
- Sanglard, D. Emerging threats in antifungal-resistant fungal pathogens. Front. Med. 2016, 3, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanbang, Y. Monitoring of Cockroaches (Orthoptera: Blattidae) Population in Bangkok Urban Area and Effective Used of Insecticides. Ph.D. Thesis, Kasetsart University, Bangkok, Thailand, 1997. [Google Scholar]
- Choate, P.; Burns, S.; Olsen, L.; Richman, D.; Perez, O.; Patnaude, M.; McFarland, C.; McManamy, K.; Pluke, R. A Dichotomous key for the identification of the cockroach fauna (Insecta: Blattaria) of Florida. Fla. Entomol. 2008, 72, 612–617. [Google Scholar]
- Buommino, E.; Vollaro, A.; Nocera, F.; Lembo, F.; DellaGreca, M.; De Martino, L.; Catania, M. Synergistic effect of abietic acid with oxacillin against methicillin-resistant Staphylococcus pseudintermedius. Antibiotics 2021, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Bates, M.K.; Phillips, D.S.; O’Bryan, J. Shaker agitation rate and orbit affect growth of cultured bacteria. Thermo Fish. Sci. Anshorbgrt 2016, 816. [Google Scholar]
- Bertino-Grimaldi, D.; Medeiros, M.N.; Vieira, R.P.; Cardoso, A.M.; Turque, A.S.; Silveira, C.B.; Martins, O.B. Bacterial community composition shifts in the gut of Periplaneta americana fed on different lignocellulosic materials. Springerplus 2013, 2, 609. [Google Scholar] [CrossRef] [Green Version]
- Bhat, K.G.; Nalawade, T.M. Antimicrobial activity of endodontic medicaments and vehicles using agar well diffusion method on facultative and obligate anaerobes. Int. J. Clin. Pediatr. Dent. 2016, 9, 335–341. [Google Scholar] [CrossRef]
- Amin, B. Isolation and characterization of antiprotozoal and antimicrobial metabolite from Penicillium roqueforti. Afr. J. Mycol. Biotech. 2016, 21, 13–26. [Google Scholar]
- Amin, B.H.; Abou-Dobara, M.I.; Diab, M.A.; Gomaa, E.A.; El-Mogazy, M.A.; El-Sonbati, A.Z.; El-Ghareib, M.S.; Hussien, M.A.; Salama, H.M. Synthesis, characterization, and biological investigation of new mixed-ligand complexes. Appl. Organomet. Chem. 2020, 34, 5689. [Google Scholar] [CrossRef]
- Nikaido, H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arias, C.A.; Murray, B.E. Antibiotic-resistant bugs in the 21st Century—A clinical super-challenge. N. Engl. J. Med. 2009, 360, 439–443. [Google Scholar] [CrossRef] [Green Version]
- Murray, C.J.; Lopez, A.D. Global and regional cause-of-death patterns in 1990. Bull. World Health Organ. 1994, 72, 447–480. [Google Scholar]
- Davies, S.C.; Fowler, T.; Watson, J.; Livermore, D.M.; Walker, D. Annual report of the chief medical officer: Infection and the rise of antimicrobial resistance. Lancet 2013, 381, 1606–1609. [Google Scholar] [CrossRef]
- Sheridan, C. Antibiotics au naturel. Nat. Biotechnol. 2006, 24, 1494–1496. [Google Scholar] [CrossRef]
- Silver, L.; Bostian, K. Screening of natural products for antimicrobial agents. Eur. J. Clin. Microbiol. Infect. Dis. 1990, 9, 455–461. [Google Scholar] [CrossRef]
- Kačániová, M.; Terentjeva, M.; Žiarovská, J.; Kowalczewski, P.L. In Vitro Antagonistic Effect of gut bacteriota isolated from indigenous honey bees and essential oils against Paenibacillus larvae. Int. J. Mol. Sci. 2020, 21, 6736. [Google Scholar] [CrossRef]
- Mahmoud, D.; Amer, A.; El-Sayed, A.K.; Rady, M. Antibacterial activity of Vespa orientalis L. venom against some antibiotic resistant bacteria. Int. J. Dev. 2017, 6, 13–20. [Google Scholar] [CrossRef]
- Iqbal, J.; Siddiqui, R.; Khan, N.A. Acanthamoeba and bacteria produce antimicrobials to target their counterpart. Parasites Vectors 2014, 7, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rady, M.; Fallata, S.; El deen, N. Environmental impact assessment of solid wastes on the housefly distribution and fly—Borne diseases. Egypt. Soc. Bioch. Mol. Biol. 2007, 2, 21–28. [Google Scholar]
- Radi, M.; Merdan, B.A.; Salama, M.S.; El Sayes, S. The first Record of Escherichia coli O157:H7 isolated from wastes and as-sociated houseflies in Egypt. World Appl. Sci. J. 2014, 31, 1437–1445. [Google Scholar]
- Fawzia, A.E.; Ashraf, A.E.; Samia, A.H.; Hend, A.E.; Nora, F.S. β-sitosterol ameliorates the chemical constituents of sunflower (Helianthus annuus L.) plants, grown under saline condition. IOSR J. Pharm. Biol. Sci. 2016, 11, 36–45. [Google Scholar]
- Douglas, A.E. Multi organismal insects: Diversity and function of resident microorganisms. Ann. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debette, J.; Blondeau, R. Présence de Pseudomonas maltophilia dans la rhizosphère de quelques plantes cultivées. Can. J. Microbiol. 1980, 26, 460–463. (In French) [Google Scholar] [CrossRef]
- Berg, R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996, 4, 430–435. [Google Scholar] [CrossRef]
- Wenke, K.; Kopka, J.; Schwachtje, J.; van Dongen, J.T.; Piechulla, B. Volatiles of rhizobacteria Serratia and Stenotrophomonas alter growth and metabolite composition of Arabidopsis thaliana. Plant Biol. 2019, 21, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Newell, P.D.; Douglas, A.E. Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 2013, 80, 788–796. [Google Scholar] [CrossRef] [Green Version]
- Savadogo, A.; Ouattara, A.C.; Bassole, H.I.; Traore, S.A. Bacteriocins and lactic acid bacteria—A mini review. Afr. J. Biotechnol. 2006, 5, 678–683. [Google Scholar]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for emerging pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; Groll, A.; Hiemenz, J.; Fleming, R.; Roilides, E.; Anaissie, E. Infections due to emerging and uncommon medically important fungal pathogens. Clin. Microbiol. Infect. 2004, 10, 48–66. [Google Scholar] [CrossRef] [Green Version]
- Gautam, A.K.; Sharma, S.; Avasthi, S.; Bhadauria, R. Diversity, pathogenicity and toxicology of A. niger: An important spoilage fungi. Res. J. Microbiol. 2011, 6, 270–280. [Google Scholar] [CrossRef]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Prim. 2018, 4, 18026. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Sun, L.; Dong, X.; Cai, Z.; Sun, X.; Yang, H.; Wang, Y.; Song, W. Characterization of a novel plant growth-promoting bacteria strain Delftia tsuruhatensis HR4 both as a diazotroph and a potential biocontrol agent against various plant pathogens. Syst. Appl. Microbiol. 2005, 28, 66–76. [Google Scholar] [CrossRef]
- Sindhu, S.S.; Rakshiya, Y.S.; Malik, D.K. Rhizosphere bacteria and their role in biological control of plant diseases. In Biotechnol Emerging Trends; Sayyed, R.Z., Patil, A.S., Eds.; Scientific Publishers: Jodhpur, India, 2009; pp. 17–52. [Google Scholar]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef]
- Etminani, F.; Harighi, B. Isolation and identification of endophytic bacteria with plant growth promoting activity and bio-control potential from wild pistachio trees. Plant Pathol. J. 2018, 34, 208. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, D.; Nazari, T.F.; Kassim, J.; Lim, S.-H. Prodigiosin—An antibacterial red pigment produced by Serratia marcescens IBRL USM 84 associated with a marine sponge Xestospongia testudinaria. J. Appl. Pharm. Sci. 2014, 4, 1–6. [Google Scholar] [CrossRef]








| Environmental Sites | Identification of Gut Bacteria | Frequency of Occurrence |
|---|---|---|
| Paper Factory | Bacillus sphaericus * | 25% |
| Ochrabactrum anthropi | 8% | |
| Stenotrophomonas maltophilia | 4% | |
| Food store | Serratia marsescens ** | 15% |
| Delftia acidovorans | 2% | |
| Kocuria rosea | 3% | |
| Sewage Water | Bacillus cereus | 4% |
| Enterobacter hormaechei | 8% | |
| Bacillus subtilis | 2% | |
| Bacillus sp. altitudinis | 20% | |
| Bacillus licheniformis | 3% | |
| Bacillus sphaericus * | - | |
| Staphylococcus aureus | 2% | |
| Escherichia coli | 2% | |
| Serratia marcescens ** | - | |
| Klebsiella pneumonia | 2% | |
| Total no. of bacterial isolates = 100 | 100% | |
| Isolated Gut Bacteria | Growth Inhibition Zone (mm) | ||||
|---|---|---|---|---|---|
| Pathogenic Bacteria | Streptococcus mutans. | Enterobacter cloacae | MRSA | Salmonella enterica | |
| Bacillus sphaericus | - | - | - | - | |
| Ochrabacterum anthropi | - | - | - | - | |
| Stenotrophomonas maltophilia | 37 ± 0.3 | - | - | - | |
| Serratia marcescens | - | - | - | - | |
| Delftia acidovorans | - | - | 12 ± 0.5 | - | |
| Kocuria rosea | - | - | 18 ± 0.4 | - | |
| Bacillus cereus | 20 ± 0.5 | - | - | - | |
| Enterobacter hormaechei | - | - | - | 8 ± 0.3 | |
| Bacillus subtilis | - | - | - | - | |
| Bacillus sp. altitudinis | - | - | 12 ± 0.3 | - | |
| Bacillus licheniformis | - | - | - | - | |
| Bacillus sphaericus | 15 ± 0.3 | - | 12 ± 0.3 | 8 ± 0.2 | |
| Staphylococcus aureus | - | 10 ±0.1 | - | - | |
| Escherichia coli | - | - | 11 ± 0.2 | - | |
| Serratia marcescens | 35 ± 0.1 | 20 ± 0.2 | 30 ± 0.1 | - | |
| Klebsiella pneumonia | - | 12 ± 0.6 | - | 8 ± 0.3 | |
| Isolated Gut Bacteria | Growth Inhibition Zone (mm) | |||||
|---|---|---|---|---|---|---|
| Pathogenic Fungi | A. niger | A. fumigatus | C. albicans | P. iticulum | A. flavus | |
| Bacillus sphaericus | - | - | - | - | - | |
| Ochrabacterium anthropi | - | - | - | - | - | |
| Stenotrophomonas sp. | - | - | - | - | - | |
| Serratia marcescens | 13 ± 0.4 | - | 11 ± 0.5 | - | - | |
| Delftia acidovorans | - | 20 ± 0.4 | - | 23 ± 0.5 | - | |
| Kocuria rosea | - | - | - | - | - | |
| Bacillus cereus | - | - | 25 ± 0.4 | - | - | |
| Enterobacter hormaechei | - | - | - | - | - | |
| Bacillus subtilis | - | 20 ± 0.3 | - | - | - | |
| Bacillus sp. altitudinis | - | - | 10 ± 0.4 | - | - | |
| Bacillus licheniformis | - | - | 12 ± 0.3 | - | - | |
| Bacillus sphaericus | - | - | - | - | - | |
| Staphylococcus aureus | - | - | - | - | - | |
| Escherichia coli | - | - | - | - | - | |
| Serratia marcescens | - | - | - | - | - | |
| Klebsiella pneumonia | - | - | - | - | 12 ± 0.4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amer, A.; Hamdy, B.; Mahmoud, D.; Elanany, M.; Rady, M.; Alahmadi, T.; Alharbi, S.; AlAshaal, S. Antagonistic Activity of Bacteria Isolated from the Periplaneta americana L. Gut against Some Multidrug-Resistant Human Pathogens. Antibiotics 2021, 10, 294. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10030294
Amer A, Hamdy B, Mahmoud D, Elanany M, Rady M, Alahmadi T, Alharbi S, AlAshaal S. Antagonistic Activity of Bacteria Isolated from the Periplaneta americana L. Gut against Some Multidrug-Resistant Human Pathogens. Antibiotics. 2021; 10(3):294. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10030294
Chicago/Turabian StyleAmer, Asmaa, Basma Hamdy, Dalia Mahmoud, Mervat Elanany, Magda Rady, Tahani Alahmadi, Sulaiman Alharbi, and Sara AlAshaal. 2021. "Antagonistic Activity of Bacteria Isolated from the Periplaneta americana L. Gut against Some Multidrug-Resistant Human Pathogens" Antibiotics 10, no. 3: 294. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10030294