Multimodal Interventions to Prevent and Control Carbapenem-Resistant Enterobacteriaceae and Extended-Spectrum β-Lactamase Producer-Associated Infections at a Tertiary Care Hospital in Egypt
Abstract
:1. Introduction
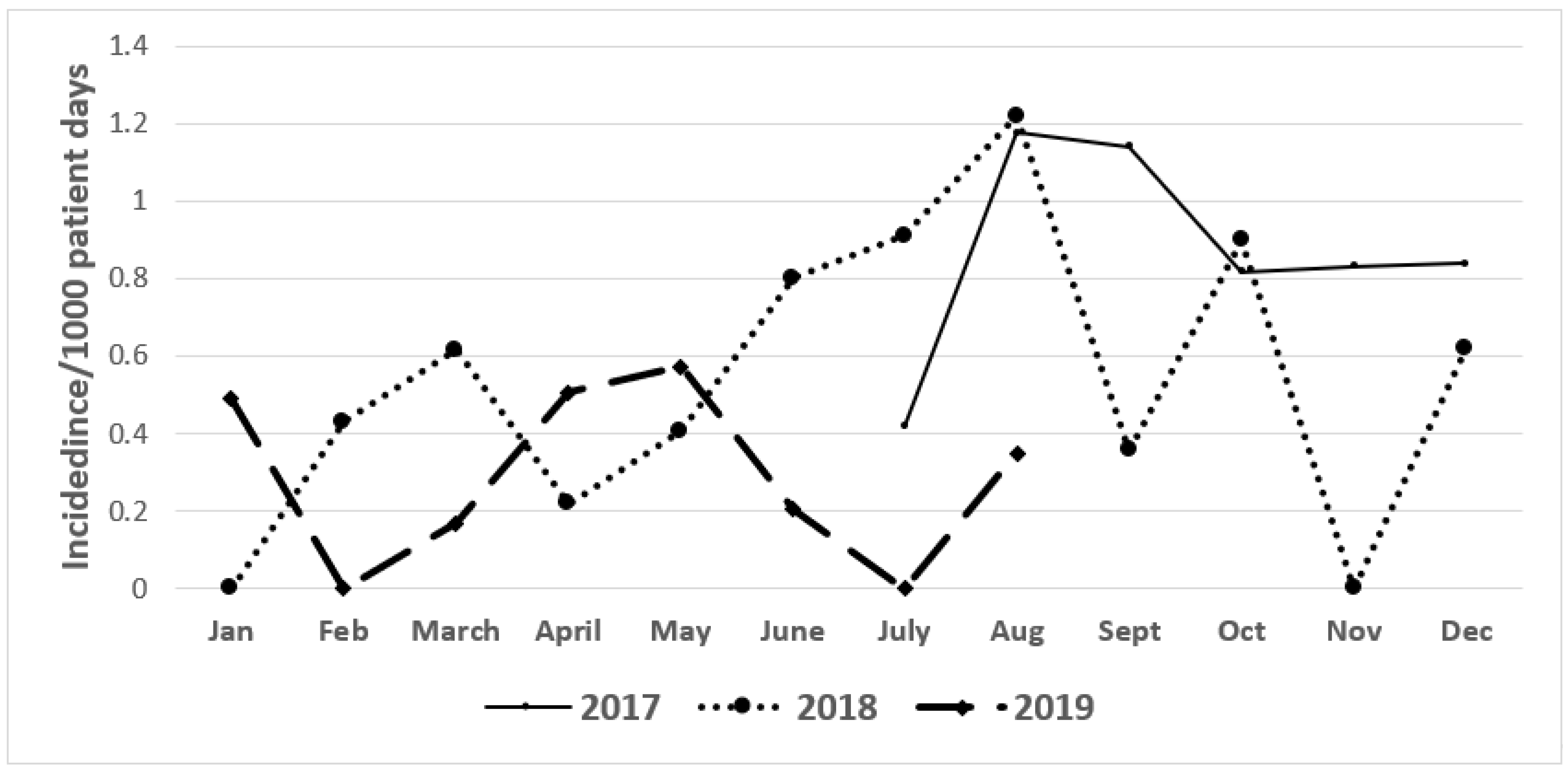
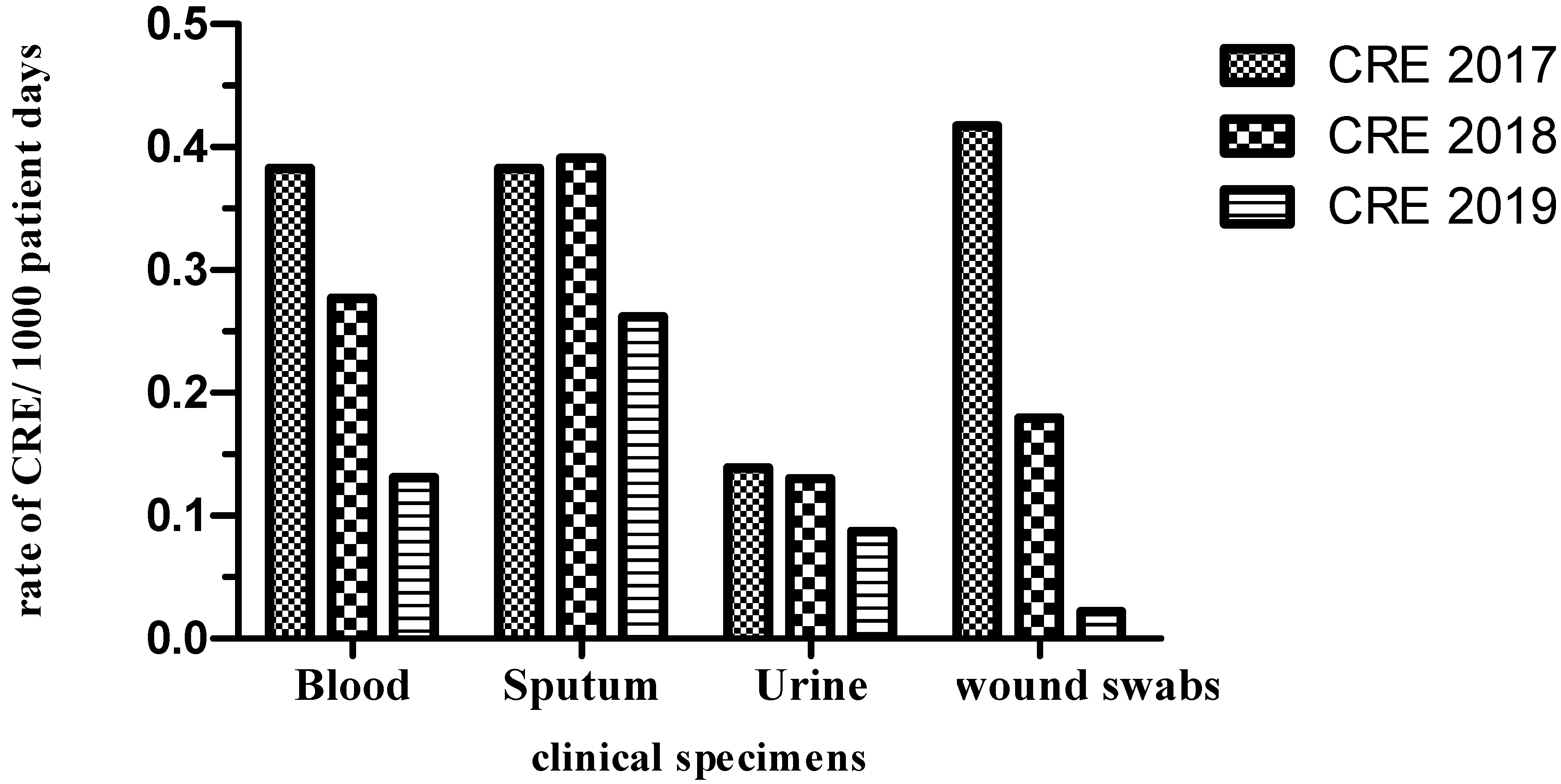
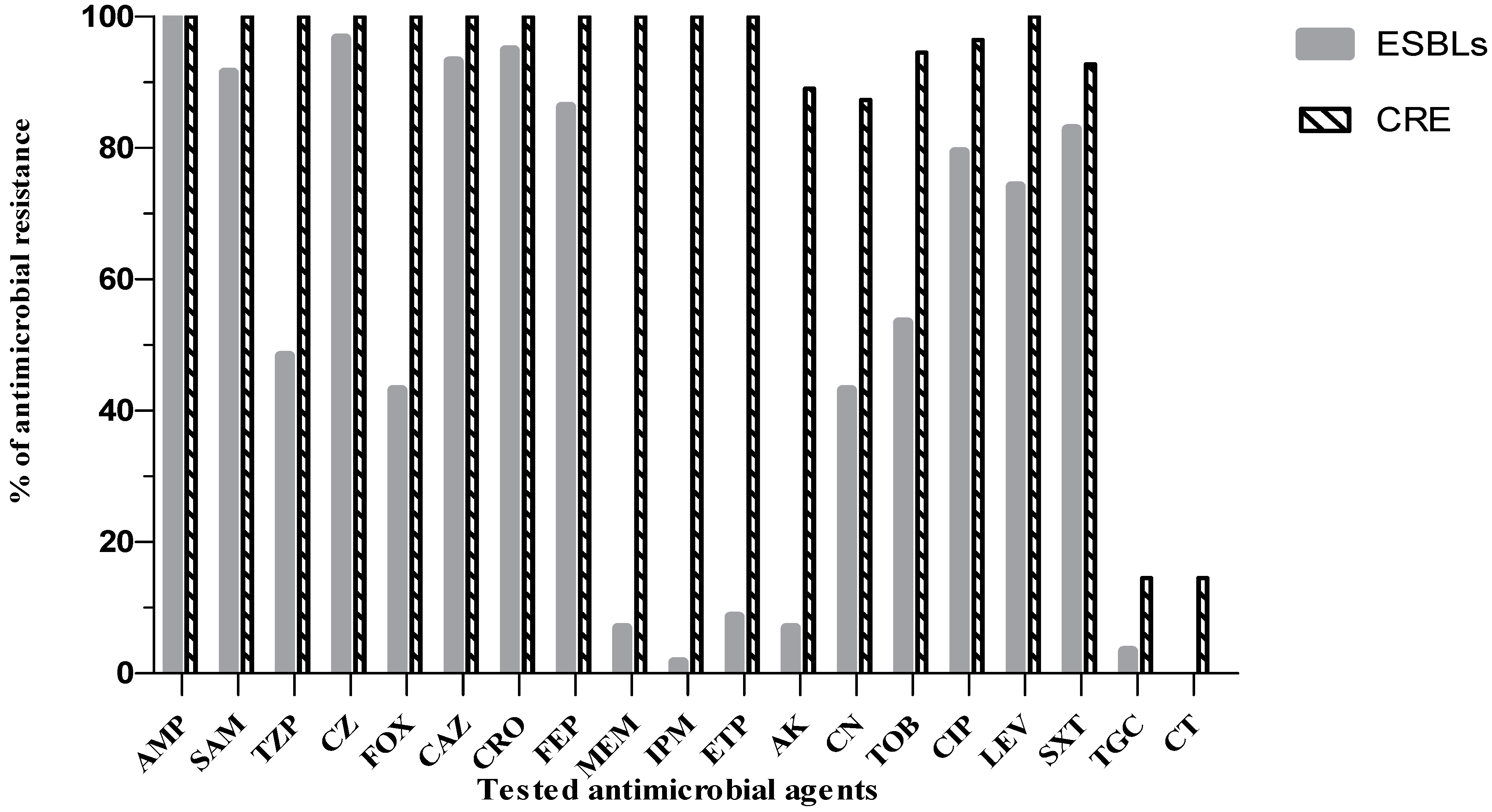
2. Results
2.1. Incidence of CRE and ESBL Producers (2017–2019)
2.2. Antibiogram Analysis of CRE and ESBL Producers
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Microbiological Procedures and Definitions
4.3. Molecular Identification of ESBL and CR Genes
4.3.1. Extraction of Chromosomal DNA from CRE and ESBL Producers
4.3.2. PCR Detection of ESBL and CR Genes
4.3.3. Sequencing of Selected PCR Products
4.4. Infection Control and Preventitive Interventions
4.4.1. Contact Isolation
4.4.2. Environmental Cleaning
4.4.3. Chlorhexidine Bathing
4.4.4. Antimicrobial Stewardship Programs
4.4.5. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Podschun, R.; Ullmann, U. Klebsiella Spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [Green Version]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella Pneumoniae: A Major Worldwide Source and Shuttle for Antibiotic Resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Mil-Homens, D.; Martins, M.; Barbosa, J.; Serafim, G.; Sarmento, M.J.; Pires, R.F.; Rodrigues, V.; Bonifácio, V.D.B.; Pinto, S.N. Carbapenem-Resistant Klebsiella Pneumoniae Clinical Isolates: In Vivo Virulence Assessment in Galleria Mellonella and Potential Therapeutics by Polycationic Oligoethyleneimine. Antibiotics 2021, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab, R.; Yasir, M.; Godfrey, R.E.; Christie, G.S.; Element, S.J.; Saville, F.; Hassan, E.A.; Ahmed, E.H.; Abu-Faddan, N.H.; Daef, E.A.; et al. Antimicrobial Resistance and Gene Regulation in Enteroaggregative Escherichia coli from Egyptian Children with Diarrhoea: Similarities and Differences. Virulence 2021, 12, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to Manage Pseudomonas Aeruginosa Infections. Drugs Context 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [Green Version]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic Resistance: A Rundown of a Global Crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [Green Version]
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T.; Mondal, A.H.; Haq, Q.M.R. Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Tacconelli, E.; Cataldo, M.A.; Dancer, S.J.; De Angelis, G.; Falcone, M.; Frank, U.; Kahlmeter, G.; Pan, A.; Petrosillo, N.; Rodríguez-Baño, J.; et al. European Society of Clinical Microbiology. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin. Microbiol. Infect. 2014, 20, 1–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redondo-Salvo, S.; Fernández-López, R.; Ruiz, R.; Vielva, L.; de Toro, M.; Rocha, E.P.C.; Garcillán-Barcia, M.P.; de la Cruz, F. Pathways for Horizontal Gene Transfer in Bacteria Revealed by a Global Map of Their Plasmids. Nat. Commun. 2020, 11, 3602. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Talaat, M.; El-Shokry, M.; El-Kholy, J.; Ismail, G.; Kotb, S.; Hafez, S.; Attia, E.; Lessa, F.C. National surveillance of health care-associated infections in Egypt: Developing a sustainable program in a resource-limited country. Am. J. Infect. Control 2016, 44, 1296–1301. [Google Scholar] [CrossRef] [Green Version]
- Abushaheen, M.A.; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; Vellappally, S.; et al. Antimicrobial Resistance, Mechanisms and Its Clinical Significance. Dis. Mon. 2020, 66, 100971. [Google Scholar] [CrossRef]
- Du, D.; Wang-Kan, X.; Neuberger, A.; Veen, H.W.; van Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug Efflux Pumps: Structure, Function and Regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Schweizer, H.P. Bacterial Resistance to Antibiotics: Active Efflux and Reduced Uptake. Adv. Drug Deliv. Rev. 2005, 57, 1486–1513. [Google Scholar] [CrossRef] [PubMed]
- Alibert, S.; N’gompaza Diarra, J.; Hernandez, J.; Stutzmann, A.; Fouad, M.; Boyer, G.; Pagès, J.-M. Multidrug Efflux Pumps and Their Role in Antibiotic and Antiseptic Resistance: A Pharmacodynamic Perspective. Expert Opin. Drug Metab. Toxicol. 2017, 13, 301–309. [Google Scholar] [CrossRef]
- King, D.T.; Sobhanifar, S.; Strynadka, N.C.J. The Mechanisms of Resistance to β-Lactam Antibiotics. In Handbook of Antimicrobial Resistance; Springer: New York, NY, USA, 2017; pp. 177–201. [Google Scholar]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Mojica, M.F.; Bonomo, R.A.; Fast, W. B1-Metallo-Beta-Lactamases: Where Do We Stand? Curr. Drug Targets 2016, 17, 1029–1050. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Interplay between β-Lactamases and New β-Lactamase Inhibitors. Nat. Rev. Microbiol. 2019, 17, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Kotra, L.P.; Haddad, J.; Mobashery, S. Aminoglycosides: Perspectives on Mechanisms of Action and Resistance and Strategies to Counter Resistance. Antimicrob. Agents Chemother. 2000, 44, 3249–3256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, M.S.; Tolmasky, M.E. Amikacin: Uses, Resistance, and Prospects for Inhibition. Molecules 2017, 22, 2267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.H.; Robicsek, A.; Jacoby, G.A.; Sahm, D.; Hooper, D.C. Prevalence in the United States of Aac(6′)-Ib-Cr Encoding a Ciprofloxacin-Modifying Enzyme. AAC 2006, 50, 3953–3955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 10 April 2021).
- Duan, N.; Du, J.; Huang, C.; Li, H. Microbial Distribution and Antibiotic Susceptibility of Lower Respiratory Tract Infections Patients From Pediatric Ward, Adult Respiratory Ward, and Respiratory Intensive Care Unit. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.A.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Age-Sex-Specific Mortality for 282 Causes of Death in 195 Countries and Territories, 1980–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef] [Green Version]
- Troeger, C.; Blacker, B.; Khalil, I.A.; Rao, P.C.; Cao, J.; Zimsen, S.R.M.; Albertson, S.B.; Deshpande, A.; Farag, T.; Abebe, Z.; et al. Estimates of the Global, Regional, and National Morbidity, Mortality, and Aetiologies of Lower Respiratory Infections in 195 Countries, 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1191–1210. [Google Scholar] [CrossRef] [Green Version]
- Jit, M.; Ng, D.H.L.; Luangasanatip, N.; Sandmann, F.; Atkins, K.E.; Robotham, J.V.; Pouwels, K.B. Quantifying the Economic Cost of Antibiotic Resistance and the Impact of Related Interventions: Rapid Methodological Review, Conceptual Framework and Recommendations for Future Studies. BMC Med. 2020, 18. [Google Scholar] [CrossRef] [Green Version]
- Ventola, C.L. The Antibiotic Resistance Crisis. P T 2015, 40, 277–283. [Google Scholar]
- Li, B.; Webster, T.J. Bacteria Antibiotic Resistance: New Challenges and Opportunities for Implant-Associated Orthopaedic Infections. J. Orthop. Res. 2018, 36, 22. [Google Scholar] [CrossRef] [Green Version]
- Center for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States, 2013; 2013. Available online: https://www.cdc.gov/ (accessed on 10 April 2021).
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage Therapy: A Potential Solution for the Antibiotic Resistance Crisis. J. Infect. Dev. Ctries 2014, 8, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Center for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States, 2019; 2019. Available online: https://www.cdc.gov/ (accessed on 10 April 2021).
- Siwakoti, S.; Subedi, A.; Sharma, A.; Baral, R.; Bhattarai, N.R.; Khanal, B. Incidence and Outcomes of Multidrug-Resistant Gram-Negative Bacteria Infections in Intensive Care Unit from Nepal- a Prospective Cohort Study. Antimicrob. Resist. Infect. Control. 2018, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sokkary, R.H.; Ramadan, R.A.; El-Shabrawy, M.; El-Korashi, L.A.; Elhawary, A.; Embarak, S.; Tash, R.M.E.; Elantouny, N.G. Community Acquired Pneumonia among Adult Patients at an Egyptian University Hospital: Bacterial Etiology, Susceptibility Profile and Evaluation of the Response to Initial Empiric Antibiotic Therapy. Infect. Drug Resist. 2018, 11, 2141–2150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadesse, B.T.; Ashley, E.A.; Ongarello, S.; Havumaki, J.; Wijegoonewardena, M.; González, I.J.; Dittrich, S. Antimicrobial Resistance in Africa: A Systematic Review. BMC Infect. Dis. 2017, 17, 616. [Google Scholar] [CrossRef]
- Singh, S.; Sharma, A.; Nag, V.L. Bacterial Pathogens from Lower Respiratory Tract Infections: A Study from Western Rajasthan. J. Fam. Med. Prim. Care 2020, 9, 1407. [Google Scholar] [CrossRef]
- Vijay, S.; Dalela, G. Prevalence of LRTI in Patients Presenting with Productive Cough and Their Antibiotic Resistance Pattern. J. Clin. Diagn. Res. 2016, 10, DC09–DC12. [Google Scholar] [CrossRef]
- Kang, H.; Zheng, W.; Kong, Z.; Jiang, F.; Gu, B.; Ma, P.; Ma, X. Disease Burden and Molecular Epidemiology of Carbapenem-Resistant Klebsiella Pneumonia Infection in a Tertiary Hospital in China. Ann. Transl. Med. 2020, 8. [Google Scholar] [CrossRef]
- Tian, D.; Pan, F.; Wang, C.; Sun, Y.; Zhang, H. Resistance Phenotype and Clinical Molecular Epidemiology of Carbapenem-Resistant Klebsiella Pneumoniae among Pediatric Patients in Shanghai. Infect. Drug Resist. 2018, 11, 1935–1943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Doyle, D.; Peirano, G.; Lascols, C.; Lloyd, T.; Church, D.L.; Pitout, J.D.D. Laboratory Detection of Enterobacteriaceae That Produce Carbapenemases. J. Clin. Microbiol. 2012, 50, 3877–3880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordmann, P.; Poirel, L.; Carrer, A.; Toleman, M.A.; Walsh, T.R. How To Detect NDM-1 Producers. J. Clin. Microbiol. 2011, 49, 718–721. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodford, N.; Tierno, P.M.; Young, K.; Tysall, L.; Palepou, M.-F.I.; Ward, E.; Painter, R.E.; Suber, D.F.; Shungu, D.; Silver, L.L.; et al. Outbreak of Klebsiella pneumoniae Producing a New Carbapenem-Hydrolyzing Class A β-Lactamase, KPC-3, in a New York Medical Center. Antimicrob. Agents Chemother. 2004, 48, 4793–4799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnet, R.; Dutour, C.; Sampaio, J.; Chanal, C.; Sirot, D.; Labia, R.; De Champs, C.; Sirot, J. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→ Gly. Antimicrob. Agents Chemother. 2001, 45, 2269–2275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasheed, J.; Jay, C.; Metchock, B.; Berkowitz, F.; Weigel, L.; Crellin, J.; Steward, C.; Hill, B.; Medeiros, A.; Tenover, F.J.A.A.; et al. Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 1997, 41, 647–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Ishikawa, J. FramePlot: A new implementation of the Frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 1999, 174, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Q.; Yin, Y.; Chen, H.; Jin, L.; Gu, B.; Xie, L.; Yang, C.; Ma, X.; Li, H.; et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae Infections: Report from the China CRE Network. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Buehrle, D.J.; Shields, R.K.; Clarke, L.G.; Potoski, B.A.; Clancy, C.J.; Nguyen, M.H. Carbapenem-Resistant Pseudomonas Aeruginosa Bacteremia: Risk Factors for Mortality and Microbiologic Treatment Failure. Antimicrob. Agents Chemother. 2016, 61. [Google Scholar] [CrossRef] [Green Version]
- Gao, B.; Li, X.; Yang, F.; Chen, W.; Zhao, Y.; Bai, G.; Zhang, Z. Molecular Epidemiology and Risk Factors of Ventilator-Associated Pneumonia Infection Caused by Carbapenem-Resistant Enterobacteriaceae. Front. Pharmacol. 2019, 10, 262. [Google Scholar] [CrossRef]
- Kumudunie, W.G.M.; Wijesooriya, L.I.; Wijayasinghe, Y.S. Comparison of Four Low-Cost Carbapenemase Detection Tests and a Proposal of an Algorithm for Early Detection of Carbapenemase-Producing Enterobacteriaceae in Resource-Limited Settings. PLoS ONE 2021, 16, e0245290. [Google Scholar] [CrossRef]
- Perez, F.; El Chakhtoura, N.G.; Papp-Wallace, K.; Wilson, B.M.; Bonomo, R.A. Treatment Options for Infections Caused by Carbapenem-Resistant Enterobacteriaceae: Can We Apply “Precision Medicine” to Antimicrobial Chemotherapy? Expert Opin. Pharmacother. 2016, 17, 761–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limbago, B.M.; Rasheed, J.K.; Anderson, K.F.; Zhu, W.; Kitchel, B.; Watz, N.; Munro, S.; Gans, H.; Banaei, N.; Kallen, A.J. IMP-Producing Carbapenem-Resistant Klebsiella Pneumoniae in the United States. J. Clin. Microbiol. 2011, 49, 4239–4245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mushtaq, S.; Vickers, A.; Doumith, M.; Ellington, M.J.; Woodford, N.; Livermore, D.M. Activity of β-Lactam/Taniborbactam (VNRX-5133) Combinations against Carbapenem-Resistant Gram-Negative Bacteria. J. Antimicrob. Chemother. 2021, 76, 160–170. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer | Primer Sequence (5’ → 3’) | Expected PCR Product Size (bp) | Ta (°C) | References |
|---|---|---|---|---|---|
| blaKPC | Pf | TGTCACTGTATCGCCGTC | 1011 | 50 | [44] |
| Pr | CTCAGTGCTCTACAGAAAACC | ||||
| blaNDM | Pf | GGTTTGGCGATCTGGTTTTC | 621 | [45] | |
| Pr | CGGAATGGCTCATCACGAT | ||||
| blaVIM | Pf | TCTACATGACCGCGTCTGTC | 748 | 50 | [46] |
| Pr | TGTGCTTTGACAACGTTCGC | ||||
| blaOXA-48 | Pf | GCGTGGTTAAGGATGAACAC | 438 | [44] | |
| Pr | CATCAAGTTCAACCCAACCG | ||||
| blaIMP | Pf | CTACCGCAGCAGAGTCTTTG | 587 | 50 | [47] |
| Pr | AACCAGTTTTGCCTTACCAT | ||||
| blaCTX-m | Pf | CGCTTTGCGATGTGCAG | 550 | 47 | [48] |
| Pr | ACCGCGATATCGTTGGT | ||||
| blaTEM | Pf | ATGAGTATTCAACATTTCCG | 867 | 47 | [49] |
| Pr | CTGACAGTTACCAATGCTTA | ||||
| blaSHV | Pf | GGTTATGCGTTATATTCGCC | 867 | 47 | [49] |
| Pr | TTAGCGTTGCCAGTGCTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamel, N.A.; Elsayed, K.M.; Awad, M.F.; Aboshanab, K.M.; El Borhamy, M.I. Multimodal Interventions to Prevent and Control Carbapenem-Resistant Enterobacteriaceae and Extended-Spectrum β-Lactamase Producer-Associated Infections at a Tertiary Care Hospital in Egypt. Antibiotics 2021, 10, 509. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10050509
Kamel NA, Elsayed KM, Awad MF, Aboshanab KM, El Borhamy MI. Multimodal Interventions to Prevent and Control Carbapenem-Resistant Enterobacteriaceae and Extended-Spectrum β-Lactamase Producer-Associated Infections at a Tertiary Care Hospital in Egypt. Antibiotics. 2021; 10(5):509. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10050509
Chicago/Turabian StyleKamel, Noha A., Khaled M. Elsayed, Mohamed F. Awad, Khaled M. Aboshanab, and Mervat I. El Borhamy. 2021. "Multimodal Interventions to Prevent and Control Carbapenem-Resistant Enterobacteriaceae and Extended-Spectrum β-Lactamase Producer-Associated Infections at a Tertiary Care Hospital in Egypt" Antibiotics 10, no. 5: 509. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10050509








