The Synergistic Effect of Biosynthesized Silver Nanoparticles and Phage ZCSE2 as a Novel Approach to Combat Multidrug-Resistant Salmonella enterica
Abstract
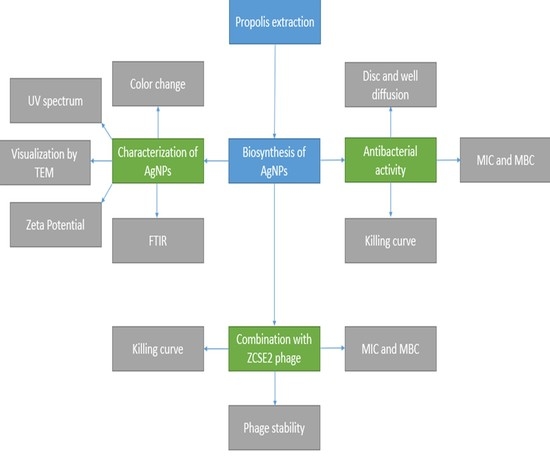
:1. Introduction
2. Results and Discussion
2.1. Characterization of AgNPs
2.2. The UV–Vis Spectrum
2.3. Visualization by TEM
2.4. Zeta Potential
2.5. FTIR Analysis
2.6. Disc and Well Diffusion Methods
2.7. Phage Stability
2.8. MIC and MBC
2.9. Time-Killing Curve
3. Materials and Methods
3.1. Preparation of Propolis
3.2. Biosynthesis of AgNPs
3.3. Characterization of AgNPs
3.3.1. UV–Vis Spectroscopy
3.3.2. FTIR Analysis
3.3.3. TEM and Zeta Potential
3.4. Antibacterial Effect of AgNPs
3.4.1. Bacterial Culture
3.4.2. MIC and MBC of AgNps
3.4.3. Antibacterial Effect of AgNPs Using Disk and Well Diffusion and Direct Spotting
3.5. Phage Combination with AgNPs
3.5.1. Phage Stability with AgNPs
3.5.2. MIC for Phage and AgNPs
3.5.3. In Vitro Time-Kill Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fashae, K.; Ogunsola, F.; Aarestrup, F.M.; Hendriksen, R.S. Antimicrobial susceptibility and serovars of Salmonella from chickens and humans in Ibadan, Nigeria. J. Infect. Dev. Ctries. 2010, 4, 484–494. [Google Scholar] [CrossRef]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The global burden of nontyphoidal salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [Green Version]
- Hyeon, J.Y.; Chon, J.W.; Hwang, I.G.; Kwak, H.S.; Kim, M.S.; Kim, S.K.; Choi, I.S.; Song, C.S.; Park, C.; Seo, K.H. Prevalence, antibiotic resistance, and molecular characterization of Salmonella serovars in retail meat products. J. Food Prot. 2011, 74, 161–166. [Google Scholar] [CrossRef]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [Green Version]
- Garvey, M. Bacteriophages and the one health approach to combat multidrug resistance: Is this the way? Antibiotics 2020, 9, 414. [Google Scholar] [CrossRef]
- Mohamed, A.; Taha, O.; El-Sherif, H.M.; Connerton, P.L.; Hooton, S.P.T.; Bassim, N.D.; Connerton, I.F.; El-Shibiny, A. Bacteriophage ZCSE2 is a Potent Antimicrobial against Salmonella enterica Serovars: Ultrastructure, genomics and efficacy. Viruses 2020, 12, 424. [Google Scholar] [CrossRef] [Green Version]
- El-Shibiny, A.; Dawoud, A. Bacteriophage applications for food safety. In Biocommunication of Phages; Witzany, G., Ed.; Springer: Cham, Switzerland, 2020; pp. 463–484. ISBN 9783030458850. [Google Scholar]
- Yosef, I.; Goren, M.G.; Globus, R.; Molshanski-Mor, S.; Qimron, U. Extending the Host Range of Bacteriophage Particles for DNA Transduction. Mol. Cell 2017, 66, 721–728.e3. [Google Scholar] [CrossRef] [Green Version]
- Seijsing, J.; Sobieraj, A.M.; Keller, N.; Shen, Y.; Zinkernagel, A.S.; Loessner, M.J.; Schmelcher, M. Improved Biodistribution and Extended Serum Half-Life of a Bacteriophage Endolysin by Albumin Binding Domain Fusion. Front. Microbiol. 2018, 9, 2927. [Google Scholar] [CrossRef]
- Chanishvili, N. Bacteriophages as Therapeutic and Prophylactic Means: Summary of the Soviet and Post Soviet Experiences. Curr. Drug Deliv. 2016. [Google Scholar] [CrossRef]
- Luong, T.; Salabarria, A.C.; Roach, D.R. Phage Therapy in the Resistance Era: Where Do We Stand and Where Are We Going? Clin. Ther. 2020, 42, 1659–1680. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef] [Green Version]
- Burmeister, A.R.; Fortier, A.; Roush, C.; Lessing, A.J.; Bender, R.G.; Barahman, R.; Grant, R.; Chan, B.K.; Turner, P.E. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 11207–11216. [Google Scholar] [CrossRef]
- Abdelsattar, A.S.; Dawoud, A.; Makky, S.; Nofal, R.; Aziz, R.K.; El-Shibiny, A. Bacteriophages: From isolation to application. Curr. Pharm. Biotechnol. 2021. [Google Scholar] [CrossRef]
- Rodriguez-Gonzalez, R.A.; Leung, C.Y.; Chan, B.K.; Turner, P.E.; Weitz, J.S. Quantitative Models of Phage-Antibiotic Combination Therapy. mSystems 2020, 5. [Google Scholar] [CrossRef] [Green Version]
- Cooper, C.J.; Koonjan, S.; Nilsson, A.S. Enhancing whole phage therapy and their derived antimicrobial enzymes through complex formulation. Pharmaceuticals 2018, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-Encoded Endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef]
- Abdelsattar, A.S.; Abdelrahman, F.; Dawoud, A.; Connerton, I.F.; El-Shibiny, A. Encapsulation of E. coli phage ZCEC5 in chitosan–alginate beads as a delivery system in phage therapy. AMB Express 2019, 9, 87. [Google Scholar] [CrossRef] [Green Version]
- Sunderland, K.S.; Yang, M.; Mao, C. Phage-Enabled Nanomedicine: From Probes to Therapeutics in Precision Medicine. Angew. Chem. Int. Ed. 2017, 56, 1964–1992. [Google Scholar] [CrossRef] [Green Version]
- Hyman, P.; Denyes, J. Bacteriophages in Nanotechnology: History and Future. In Bacteriophages: Biology, Technology, Therapy; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 1–31. ISBN 9783319405988. [Google Scholar]
- Bae, E.; Park, H.-J.; Lee, J.; Kim, Y.; Yoon, J.; Park, K.; Choi, K.; Yi, J. Bacterial cytotoxicity of the silver nanoparticle related to physicochemical metrics and agglomeration properties. Environ. Toxicol. Chem. 2010, 29, 2154–2160. [Google Scholar] [CrossRef]
- Silver, S.; Phung, L.T.; Silver, G. Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J. Ind. Microbiol. Biotechnol. 2006, 33, 627–634. [Google Scholar] [CrossRef]
- Su, H.L.; Lin, S.H.; Wei, J.C.; Pao, I.C.; Chiao, S.H.; Huang, C.C.; Lin, S.Z.; Lin, J.J. Novel nanohybrids of silver particles on clay platelets for inhibiting silver-resistant bacteria. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Lara, H.H.; Ayala-Núñez, N.V.; del Turrent, L.C.I.; Padilla, C.R. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J. Microbiol. Biotechnol. 2010, 26, 615–621. [Google Scholar] [CrossRef]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, S.M.; Hess, K.L.; Gearhart, J.M.; Geiss, K.T.; Schlager, J.J. In vitro toxicity of nanoparticles in BRL 3A rat liver cells. Toxicol. In Vitro 2005, 19, 975–983. [Google Scholar] [CrossRef]
- Lam, C.W.; James, J.T.; McCluskey, R.; Hunter, R.L. Pulmonary toxicity of single-wall carbon nanotubes in mice 7 and 90 days after intractracheal instillation. Toxicol. Sci. 2004, 77, 126–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kischkel, B.; de Castilho, P.F.; de Oliveira, K.M.P.; Rezende, P.S.T.; Bruschi, M.L.; Svidzinski, T.I.E.; Negri, M. Silver nanoparticles stabilized with propolis show reduced toxicity and potential activity against fungal infections. Future Microbiol. 2020, 15, 521–539. [Google Scholar] [CrossRef]
- Ghramh, H.A.; Khan, K.A.; Ibrahim, E.H.; Ansari, M.J. Biogenic synthesis of silver nanoparticles using propolis extract, their characterization, and biological activities. Sci. Adv. Mater. 2019, 11, 876–883. [Google Scholar] [CrossRef]
- Castaldo, S.; Capasso, F. Propolis, an old remedy used in modern medicine. Fitoterapia 2002, 73, S1–S6. [Google Scholar] [CrossRef]
- Ramadan, A.; Soliman, G.; Mahmoud, S.S.; Nofal, S.M.; Abdel-Rahman, R.F. Evaluation of the safety and antioxidant activities of Crocus sativus and Propolis ethanolic extracts. J. Saudi Chem. Soc. 2012, 16, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Wu, J.; Wei, J.; Pu, L.; Guo, C.; Yang, J.; Yang, M.; Luo, H. Brazilian green propolis improves immune function in aged mice. J. Clin. Biochem. Nutr. 2014, 55, 7–10. [Google Scholar] [CrossRef] [Green Version]
- Shimabuku, Q.L.; Ueda-Nakamura, T.; Bergamasco, R.; Fagundes-Klen, M.R. Chick-Watson kinetics of virus inactivation with granular activated carbon modified with silver nanoparticles and/or copper oxide. Process Saf. Environ. Prot. 2018, 117, 33–42. [Google Scholar] [CrossRef]
- Gilcrease, E.; Williams, R.; Goel, R. Evaluating the effect of silver nanoparticles on bacteriophage lytic infection cycle-a mechanistic understanding. Water Res. 2020, 181, 115900. [Google Scholar] [CrossRef]
- Ahiwale, S.S.; Bankar, A.V.; Tagunde, S.; Kapadnis, B.P. A Bacteriophage Mediated Gold Nanoparticles Synthesis and Their Anti-biofilm Activity. Indian J. Microbiol. 2017, 57, 188–194. [Google Scholar] [CrossRef]
- Manoharadas, S.; Altaf, M.; Alrefaei, A.F.; Devasia, R.M.; Badjah Hadj, A.Y.M.; Abuhasil, M.S.A. Concerted dispersion of Staphylococcus aureus biofilm by bacteriophage and “green synthesized” silver nanoparticles. RSC Adv. 2021, 11, 1420–1429. [Google Scholar] [CrossRef]
- Jafari, A.; Vaghari, H.; Jafarizadeh-Malmiri, H. Development of Antimicrobial Films Based on Aloe vera and Fabricated AgNPs Using Propolis. Proc. Natl. Acad. Sci. USA India Sect. B Biol. Sci. 2020, 91, 95–103. [Google Scholar] [CrossRef]
- Mohammadlou, M.; Maghsoudi, H.; Jafarizadeh-Malmiri, H. A review on green silver nanoparticles based on plants: Synthesis, potential applications and eco-friendly approach. Int. Food Res. J. 2016, 23, 446–463. [Google Scholar]
- Przybyłek, I.; Karpiński, T.M. Antibacterial properties of propolis. Molecules 2019, 24, 2047. [Google Scholar] [CrossRef] [Green Version]
- Sharma, G.; Nam, J.-S.; Sharma, A.R.; Lee, S.-S. Antimicrobial potential of silver nanoparticles synthesized using medicinal herb coptidis rhizome. Molecules 2018, 23, 2268. [Google Scholar] [CrossRef] [Green Version]
- Jagtap, U.B.; Bapat, V.A. Green synthesis of silver nanoparticles using Artocarpus heterophyllus Lam. seed extract and its antibacterial activity. Ind. Crops Prod. 2013, 46, 132–137. [Google Scholar] [CrossRef]
- Ballauff, M.; Lu, Y. “Smart” nanoparticles: Preparation, characterization and applications. Polymer 2007, 48, 1815–1823. [Google Scholar] [CrossRef] [Green Version]
- Ajitha, B.; Reddy, Y.A.K.; Reddy, P.S.; Jeon, H.-J.; Ahn, C.W. Role of capping agents in controlling silver nanoparticles size, antibacterial activity and potential application as optical hydrogen peroxide sensor. RSC Adv. 2016, 6, 36171–36179. [Google Scholar] [CrossRef]
- Niska, K.; Knap, N.; Kędzia, A.; Jaskiewicz, M.; Kamysz, W.; Inkielewicz-Stepniak, I. Capping agent-dependent toxicity and antimicrobial activity of silver nanoparticles: An in vitro study. Concerns about potential application in dental practice. Int. J. Med. Sci. 2016, 13, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, F.; Iqbal, S.; Khalid, N.; Hussain, I.; Hussain, Z.; Szmigielski, R.; Janjua, H.A. Screening and stability testing of commercially applicable Heliotropium crispum silver nanoparticle formulation with control over aging and biostability. Appl. Nanosci. 2020, 10, 1941–1956. [Google Scholar] [CrossRef]
- Thiyagarajan, K.; Bharti, V.K.; Tyagi, S.; Tyagi, P.K.; Ahuja, A.; Kumar, K.; Raj, T.; Kumar, B. Synthesis of non-toxic, biocompatible, and colloidal stable silver nanoparticle using egg-white protein as capping and reducing agents for sustainable antibacterial application. RSC Adv. 2018, 8, 23213–23229. [Google Scholar] [CrossRef] [Green Version]
- Ardani, H.K.; Imawan, C.; Handayani, W.; Djuhana, D.; Harmoko, A.; Fauzia, V. Enhancement of the stability of silver nanoparticles synthesized using aqueous extract of Diospyros discolor Willd. leaves using polyvinyl alcohol. In Proceedings of the IOP Conference Series: Materials Science and Engineering, Busan, Korea, 26–28 August 2017; IOP Publishing: Bristol, UK, 2017; Volume 188, p. 12056. [Google Scholar]
- Abdelsattar, A.S.; Dawoud, A.; Helal, M.A. Interaction of nanoparticles with biological macromolecules: A review of molecular docking studies. Nanotoxicology 2020, 15, 1–30. [Google Scholar]
- Ahsan, S.M.; Rao, C.M.; Ahmad, M.F. Nanoparticle-protein interaction: The significance and role of protein corona. Cell. Mol. Toxicol. Nanopart. 2018, 1048, 175–198. [Google Scholar]
- Peng, S.; McMahon, J.M.; Schatz, G.C.; Gray, S.K.; Sun, Y. Reversing the size-dependence of surface plasmon resonances. Proc. Natl. Acad. Sci. USA 2010, 107, 14530–14534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jitian, S.; Bratu, I. Determination of optical constants of polymethyl methacrylate films from IR reflection-absorption spectra. In Proceedings of the AIP Conference Proceedings 1425, Timisoara, Romania, 16–18 April 2018; American Institute of Physics: College Park, MD, USA, 2018; Volume 2012, pp. 26–29. [Google Scholar]
- Markham, K.R.; Mitchell, K.A.; Wilkins, A.L.; Daldy, J.A.; Lu, Y. HPLC and GC-MS identification of the major organic constituents in New Zeland propolis. Phytochemistry 1996, 42, 205–211. [Google Scholar] [CrossRef]
- Rastogi, L.; Arunachalam, J. Sunlight based irradiation strategy for rapid green synthesis of highly stable silver nanoparticles using aqueous garlic (Allium sativum) extract and their antibacterial potential. Mater. Chem. Phys. 2011, 129, 558–563. [Google Scholar] [CrossRef]
- Shameli, K.; Ahmad, M.B.; Zamanian, A.; Sangpour, P.; Shabanzadeh, P.; Abdollahi, Y.; Zargar, M. Green biosynthesis of silver nanoparticles using Curcuma longa tuber powder. Int. J. Nanomed. 2012, 7, 5603. [Google Scholar] [CrossRef] [Green Version]
- Corciova, A.; Mircea, C.; Burlec, A.-F.; Cioanca, O.; Tuchilus, C.; Fifere, A.; Lungoci, A.-L.; Marangoci, N.; Hancianu, M. Antioxidant, antimicrobial and photocatalytic activities of silver nanoparticles obtained by bee propolis extract assisted biosynthesis. Farmacia 2019, 67, 482–489. [Google Scholar] [CrossRef]
- Yu, J.; Wu, P. Crystallization process of poly (ɛ-caprolactone)–poly (ethylene oxide)–poly (ɛ-caprolactone) investigated by infrared and two-dimensional infrared correlation spectroscopy. Polymer 2007, 48, 3477–3485. [Google Scholar] [CrossRef]
- Chen, J.; Li, S.; Luo, J.; Wang, R.; Ding, W. Enhancement of the antibacterial activity of silver nanoparticles against phytopathogenic bacterium Ralstonia solanacearum by stabilization. J. Nanomater. 2016. [Google Scholar] [CrossRef] [Green Version]
- Randall, C.P.; Oyama, L.B.; Bostock, J.M.; Chopra, I.; O’Neill, A.J. The silver cation (Ag+): Antistaphylococcal activity, mode of action and resistance studies. J. Antimicrob. Chemother. 2013, 68, 131–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seong, M.; Lee, D.G. Silver nanoparticles against Salmonella enterica serotype typhimurium: Role of inner membrane dysfunction. Curr. Microbiol. 2017, 74, 661–670. [Google Scholar] [CrossRef]
- Xiang, D.; Zheng, Y.; Duan, W.; Li, X.; Yin, J.; Shigdar, S.; O’Connor, M.L.; Marappan, M.; Zhao, X.; Miao, Y. Inhibition of A/Human/Hubei/3/2005 (H3N2) influenza virus infection by silver nanoparticles in vitro and in vivo. Int. J. Nanomed. 2013, 8, 4103. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.; Kaushik, S.; Pandit, P.; Dhull, D.; Yadav, J.P.; Kaushik, S. Green synthesis of silver nanoparticles from medicinal plants and evaluation of their antiviral potential against chikungunya virus. Appl. Microbiol. Biotechnol. 2019, 103, 881–891. [Google Scholar] [CrossRef]
- Omara, S.T.; Zawrah, M.F.; Samy, A.A. Minimum bactericidal concentration of chemically synthesized silver nanoparticles against pathogenic Salmonella and Shigella strains isolated from layer poultry farms. J. Appl. Pharm. Sci. 2017, 7, 214–221. [Google Scholar]
- Hamouda, R.A.; Yousuf, W.E.; Abdeen, E.E.; Mohamed, A. Biological and chemical synthesis of silver nanoparticles: Characterization, MIC and antibacterial activity against pathogenic bacteria. J. Chem. Pharm. Res 2019, 11, 1–12. [Google Scholar]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prere, M.-F.; Krisch, H.M. Phage-antibiotic synergy (PAS): β-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef] [Green Version]
- Cooper, C.J.; Khan Mirzaei, M.; Nilsson, A.S. Adapting drug approval pathways for bacteriophage-based therapeutics. Front. Microbiol. 2016, 7, 1209. [Google Scholar] [CrossRef]
- Liu, Y.-Q.; Zhang, Y.-Z.; Sun, C.-Y.; Gao, P.-J. A novel approach to estimate in vitro antibacterial potency of Chinese medicine using a concentration-killing curve method. Am. J. Chin. Med. 2005, 33, 671–682. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [Green Version]
- Hooton, S.P.T.; Atterbury, R.J.; Connerton, I.F. Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. Int. J. Food Microbiol. 2011, 151, 157–163. [Google Scholar] [CrossRef]
- Parvekar, P.; Palaskar, J.; Metgud, S.; Maria, R.; Dutta, S. The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of silver nanoparticles against Staphylococcus aureus. Biomater. Investig. Dent. 2020, 7, 105–109. [Google Scholar] [CrossRef]
- Baldi, F.; Daniele, S.; Gallo, M.; Paganelli, S.; Battistel, D.; Piccolo, O.; Faleri, C.; Puglia, A.M.; Gallo, G. Polysaccharide-based silver nanoparticles synthesized by Klebsiella oxytoca DSM 29614 cause DNA fragmentation in E. coli cells. BioMetals 2016, 29, 321–331. [Google Scholar] [CrossRef]
- Ajitha, B.; Reddy, Y.A.K.; Shameer, S.; Rajesh, K.M.; Suneetha, Y.; Reddy, P.S. Lantana camara leaf extract mediated silver nanoparticles: Antibacterial, green catalyst. J. Photochem. Photobiol. B Biol. 2015, 149, 84–92. [Google Scholar] [CrossRef]






| Serial Dilution | Dimeter (mm) | ||
|---|---|---|---|
| Direct Spotting | Disk Diffusion | Well Diffusion | |
| 3% and 1.5% Propolis extract | without antibacterial effect | 0 | 0 |
| 184 µ/mL of AgNPs | antibacterial effect | 11 | 12 |
| 92 µg/mL of AgNPs | antibacterial effect | 10 | 10 |
| 46 µg/mL of AgNPs | antibacterial effect | 8 | 10 |
| 23 µg/mL of AgNPs | antibacterial effect | 9 | 7 |
| 11.5 µg/mL of AgNPs | without antibacterial effect | 8 | 6~7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelsattar, A.S.; Nofal, R.; Makky, S.; Safwat, A.; Taha, A.; El-Shibiny, A. The Synergistic Effect of Biosynthesized Silver Nanoparticles and Phage ZCSE2 as a Novel Approach to Combat Multidrug-Resistant Salmonella enterica. Antibiotics 2021, 10, 678. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10060678
Abdelsattar AS, Nofal R, Makky S, Safwat A, Taha A, El-Shibiny A. The Synergistic Effect of Biosynthesized Silver Nanoparticles and Phage ZCSE2 as a Novel Approach to Combat Multidrug-Resistant Salmonella enterica. Antibiotics. 2021; 10(6):678. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10060678
Chicago/Turabian StyleAbdelsattar, Abdallah S., Rana Nofal, Salsabil Makky, Anan Safwat, Amera Taha, and Ayman El-Shibiny. 2021. "The Synergistic Effect of Biosynthesized Silver Nanoparticles and Phage ZCSE2 as a Novel Approach to Combat Multidrug-Resistant Salmonella enterica" Antibiotics 10, no. 6: 678. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10060678