RND Efflux Systems Contribute to Resistance and Virulence of Aliarcobacter butzleri
Abstract
:1. Introduction
2. Results and Discussion
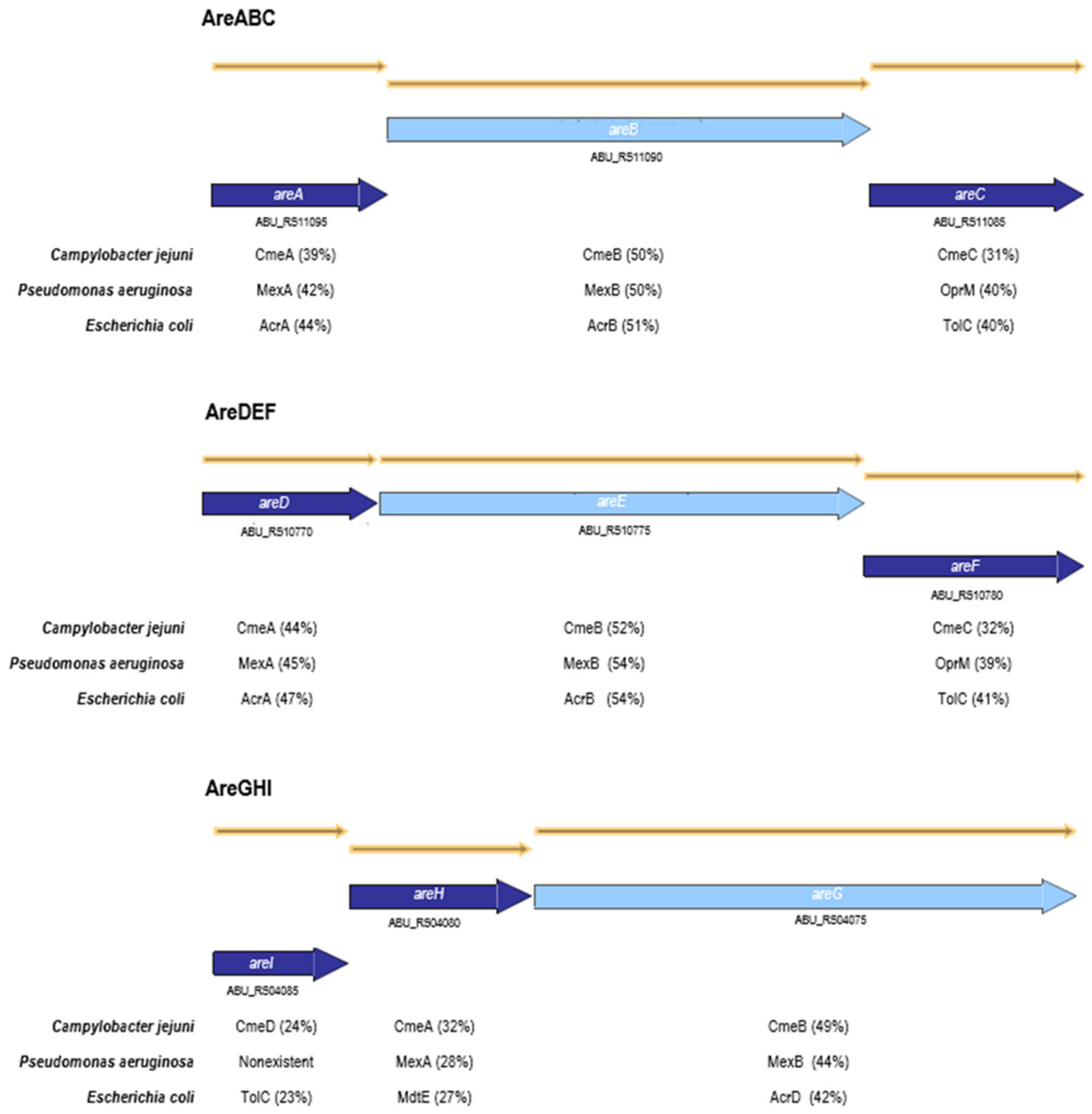
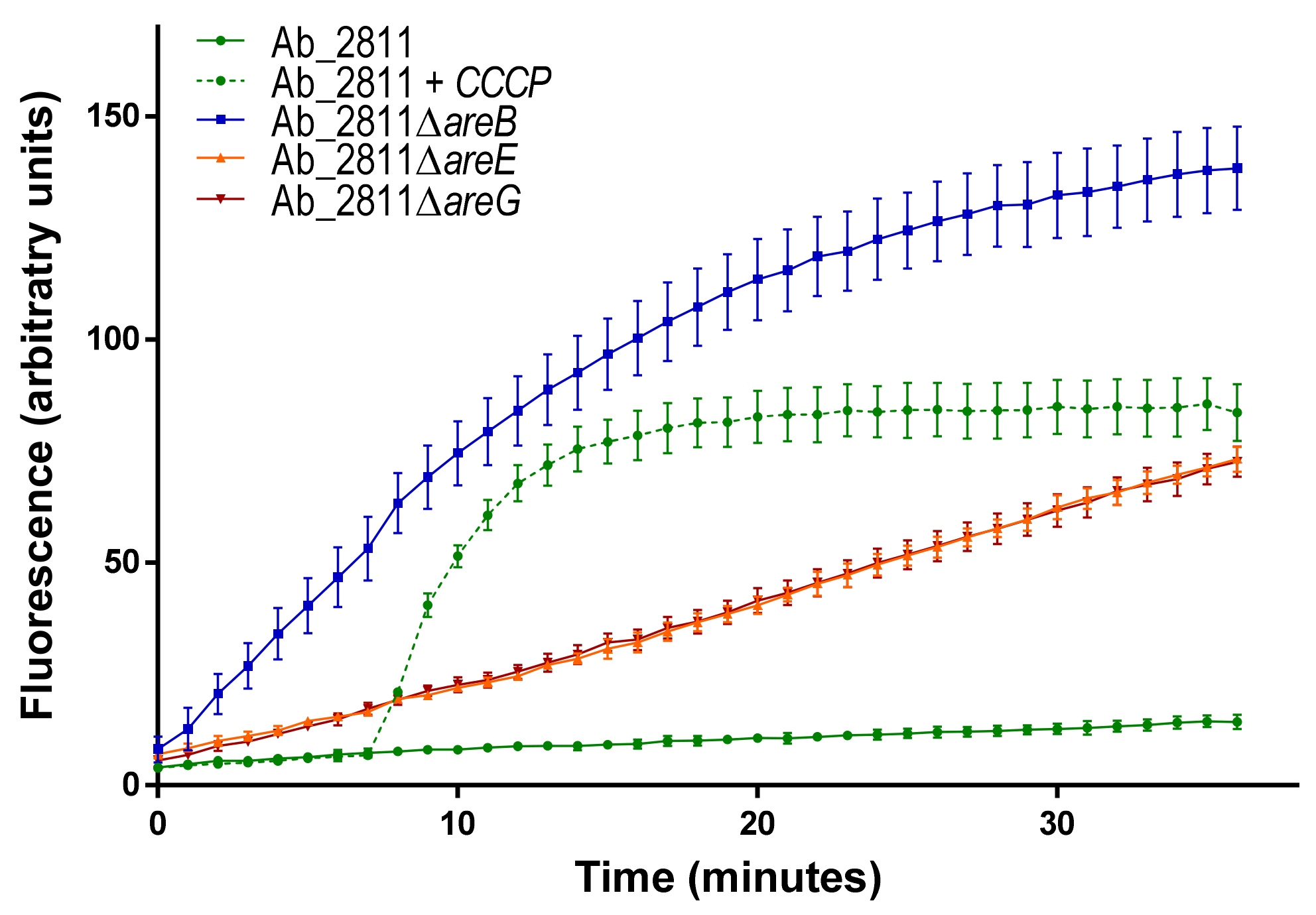
2.1. Impact of the Inactivation of Three RND Efflux Pumps on Bacterial Resistance
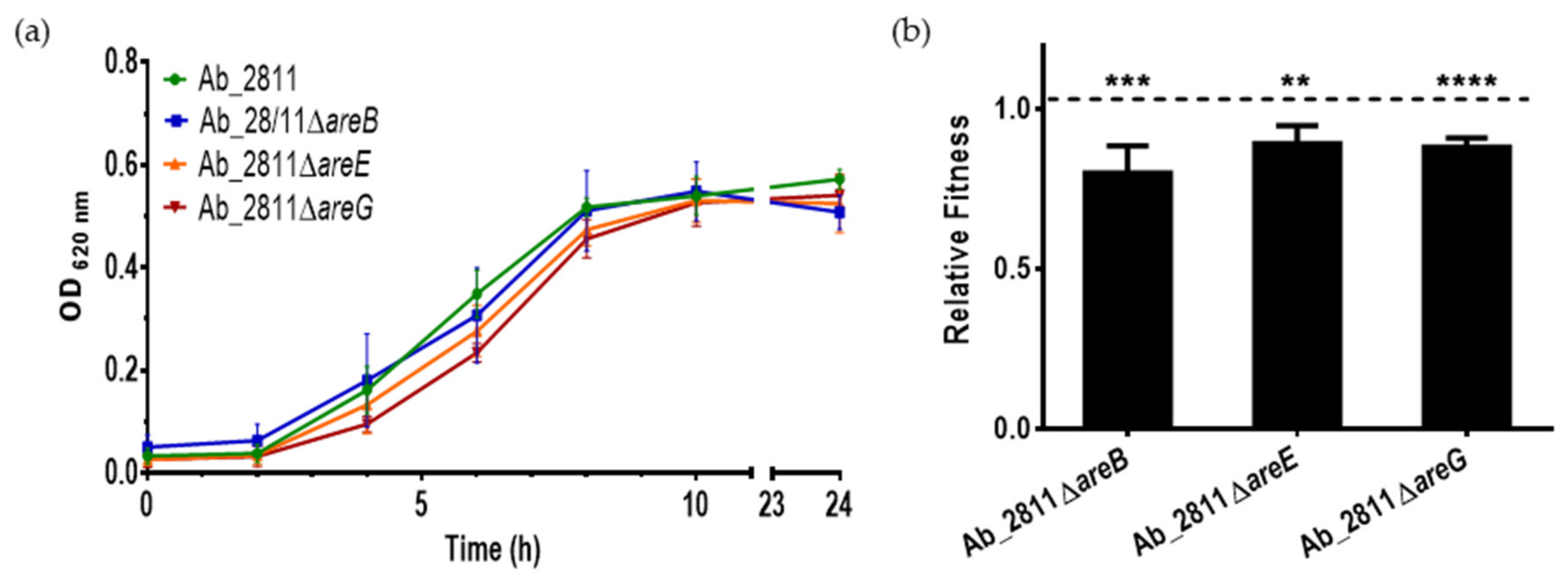
2.2. Influence of Efflux System Impairment in Bacterial Growth and Relative Fitness
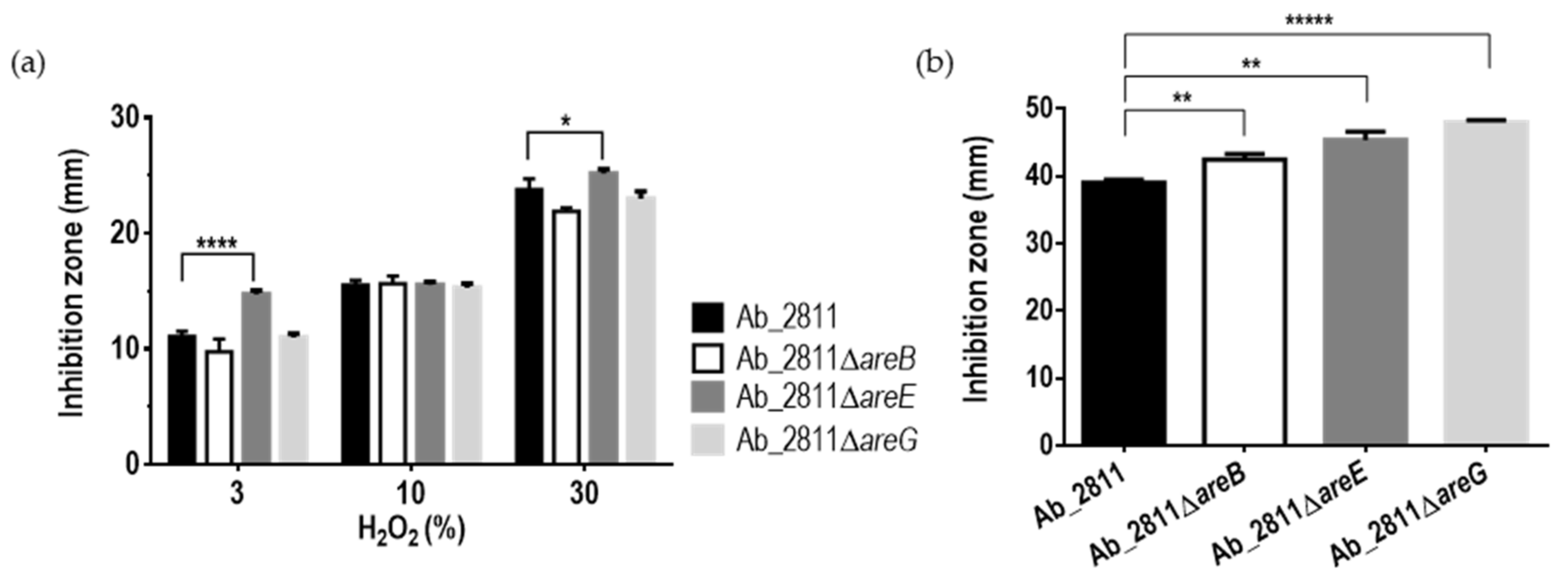
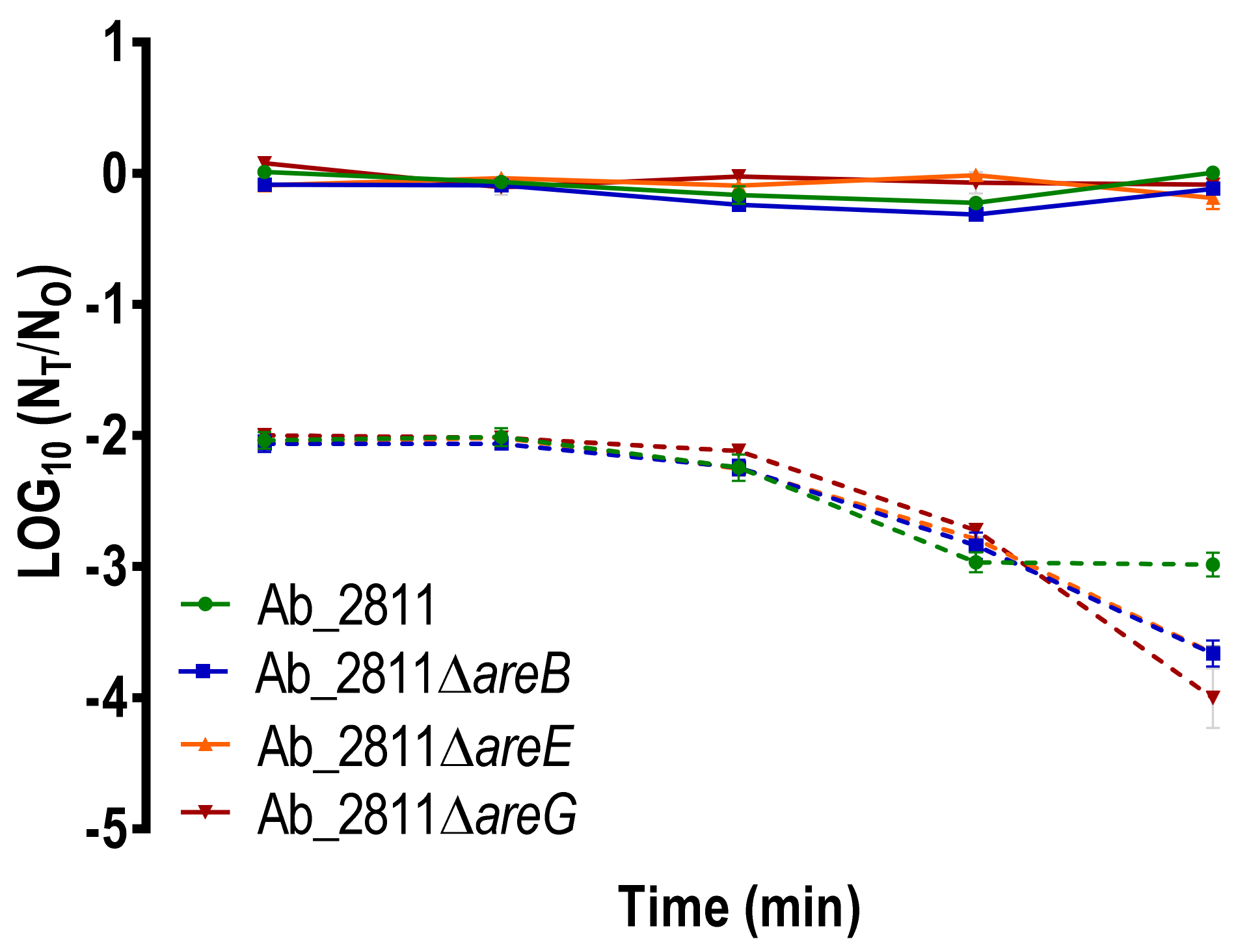
2.3. Effect of Inactivation of the RND Efflux Pumps in the Susceptibility to Oxidative and Bile Stress
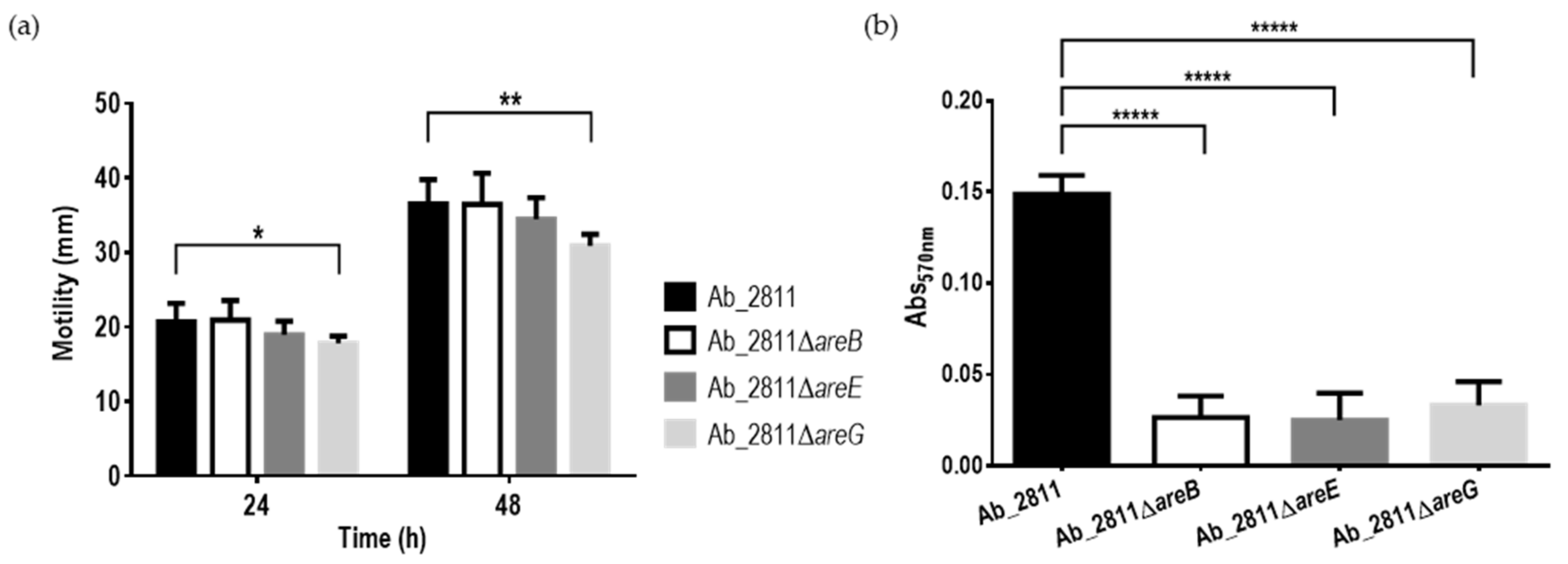
2.4. Impact of the Inactivation of RND Efflux Pumps on Motility and Biofilm Formation Ability
2.5. Impact of the Inactivation of RND Efflux Pumps on Human Serum Susceptibility
2.6. In Vitro Adhesion to and Invasion into Caco-2 Cells
3. Materials and Methods
3.1. Bacterial Strains and Growth Conditions
3.2. Construction of Aliarcobacter butzleri Efflux Pump Mutants
3.3. Antimicrobial Susceptibility Test
3.4. Ethidium Bromide Accumulation Assay
3.5. Growth Curves and Competition Experiments of the Parental and Mutant Strains
3.6. Stress Susceptibility Assays
3.7. Motility Assay
3.8. Biofilm Formation Ability
3.9. Serum Killing Assays
3.10. Adhesion and Invasion Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pérez-Cataluña, A.; Salas-Massó, N.; Diéguez, A.L.; Balboa, S.; Lema, A.; Romalde, J.L.; Figueras, M.J. Revisiting the Taxonomy of the Genus Arcobacter: Getting Order from the Chaos. Front. Microbiol. 2018, 9, 2077. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. List of New Names and New Combinations Previously Effectively, but Not Validly, Published. Int. J. Syst. Evol. Microbiol. 2020, 70, 1–5. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. List of New Names and New Combinations Previously Effectively, but not Validly, Published. Int. J. Syst. Evol. Microbiol. 2020, 70, 2960–2966. [Google Scholar] [CrossRef] [PubMed]
- Whiteduck-Léveillée, K.; Whiteduck-Léveillée, J.; Cloutier, M.; Tambong, J.T.; Xu, R.; Topp, E.; Arts, M.; Chao, J.; Adam, Z.; Lévesque, C.A.; et al. Arcobacter Lanthieri sp. nov., Isolated from Pig and Dairy Cattle Manure. Int. J. Syst. Evol. Microbiol. 2015, 65, 2709–2716. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.; Queiroz, J.; Oleastro, M.; Domingues, F.C. Insights in the Pathogenesis and Resistance of Arcobacter: A Review. Crit. Rev. Microbiol. 2015, 42, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Chieffi, D.; Fanelli, F.; Fusco, V. Arcobacter butzleri: Up-to-Date Taxonomy, Ecology, and Pathogenicity of An Emerging Pathogen. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2071–2109. [Google Scholar] [CrossRef]
- Collado, L.; Figueras, M.J. Taxonomy, Epidemiology, and Clinical Relevance of the Genus Arcobacter. Clin. Microbiol. Rev. 2011, 24, 174–192. [Google Scholar] [CrossRef] [Green Version]
- Figueras, M.J.; Levican, A.; Pujol, I.; Ballester, F.; Quilez, M.J.R.; Gomez-Bertomeu, F. A Severe Case of Persistent Diarrhoea Associated with Arcobacter cryaerophilus but Attributed to Campylobacter sp. and A Review of the Clinical Incidence of Arcobacter Spp. New Microbes New Infect. 2014, 2, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Kerkhof, P.-J.; Abeele, A.-M.V.D.; Strubbe, B.; Vogelaers, D.; Vandamme, P.; Houf, K. Diagnostic Approach for Detection and Identification of Emerging Enteric Pathogens Revisited: The (Ali)arcobacter Lanthieri Case. New Microbes New Infect. 2021, 39, 100829. [Google Scholar] [CrossRef]
- Abeele, A.-M.V.D.; Vogelaers, D.; Van Hende, J.; Houf, K. Prevalence of Arcobacter Species among Humans, Belgium, 2008–2013. Emerg. Infect. Dis. 2014, 20, 1746–1749. [Google Scholar] [CrossRef] [Green Version]
- ICMSF International Commission on Microbiological Specifications for Foods. Microorganisms in Foods 7: Microbiological Testing in Food Safety Management; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2002; p. 171. [Google Scholar]
- Abdelbaqi, K.; Mãnard, A.; Prouzet-Mauleon, V.; Bringaud, F.; Lehours, P.; Mãgraud, F. Nucleotide Sequence of the gyrA Gene of Arcobacter species and Characterization of Human Ciprofloxacin-Resistant Clinical Isolates. FEMS Immunol. Med. Microbiol. 2007, 49, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Isidro, J.; Ferreira, S.; Pinto, M.; Domingues, F.; Oleastro, M.; Gomes, J.P.; Borges, V. Virulence and Antibiotic Resistance Plasticity of Arcobacter butzleri: Insights on the Genomic Diversity of An Emerging Human Pathogen. Infect. Genet. Evol. 2020, 80, 104213. [Google Scholar] [CrossRef]
- Ferreira, S.; Silva, A.L.; Tomás, J.; Mateus, C.; Domingues, F.; Oleastro, M. Characterization of AreABC, An RND-type Efflux System Involved in Antimicrobial Resistance of Aliarcobacter butzleri. Antimicrob. Agents Chemother. 2021, 65, AAC0072921. [Google Scholar] [CrossRef]
- Blair, J.; Piddock, L.J. Structure, Function and Inhibition of RND Efflux Pumps in Gram-Negative Bacteria: An Update. Curr. Opin. Microbiol. 2009, 12, 512–519. [Google Scholar] [CrossRef]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martínez, J.L. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, K.A.; Liu, Q.; Elbourne, L.D.; Ahmad, I.; Sharples, D.; Naidu, V.; Chan, C.L.; Li, L.; Harborne, S.; Pokhrel, A.; et al. Pacing across the Membrane: The Novel PACE Family of Efflux Pumps is Widespread in Gram-Negative Pathogens. Res. Microbiol. 2018, 169, 450–454. [Google Scholar] [CrossRef]
- Blair, J.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Schweizer, H.P. Bacterial Resistance to Antibiotics: Active Efflux and Reduced Uptake. Adv. Drug Deliv. Rev. 2005, 57, 1486–1513. [Google Scholar] [CrossRef] [PubMed]
- Hirakata, Y.; Srikumar, R.; Poole, K.; Gotoh, N.; Suematsu, T.; Kohno, S.; Kamihira, S.; Hancock, R.; Speert, D.P. Multidrug Efflux Systems Play an Important Role in the Invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 2002, 196, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Sahin, O.; Michel, L.O.; Zhang, Q. Critical Role of Multidrug Efflux Pump CmeABC in Bile Resistance and In Vivo Colonization of Campylobacter jejuni. Infect. Immun. 2003, 71, 4250–4259. [Google Scholar] [CrossRef] [Green Version]
- Padilla, E.; Llobet, E.; Doménech-Sánchez, A.; Martínez-Martínez, L.; Bengoechea, J.; Albertí, S. Klebsiella pneumoniae AcrAB Efflux Pump Contributes to Antimicrobial Resistance and Virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Pérez, A.; Poza, M.; Fernández, A.; Fernández, M.D.C.; Mallo, S.; Merino, M.; Rumbo-Feal, S.; Cabral, M.P.; Bou, G. Involvement of the AcrAB-TolC Efflux Pump in the Resistance, Fitness, and Virulence of Enterobacter cloacae. Antimicrob. Agents Chemother. 2012, 56, 2084–2090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang-Kan, X.; Blair, J.M.A.; Chirullo, B.; Betts, J.; La Ragione, R.M.; Ivens, A.; Ricci, V.; Opperman, T.J.; Piddock, L.J.V. Lack of AcrB Efflux Function Confers Loss of Virulence on Salmonella enterica Serovar Typhimurium. mBio 2017, 8, e00968-17. [Google Scholar] [CrossRef] [Green Version]
- Nikaido, H. Structure and Mechanism of RND-Type Multidrug Efflux Pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 2011, 77, 1–60. [Google Scholar]
- Nikaido, H.; Takatsuka, Y. Mechanisms of RND Multidrug Efflux Pumps. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 769–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ealvarez-Ortega, C.; Eolivares, J.; Martínez, J.L. RND Multidrug Efflux Pumps: What are They Good for? Front. Microbiol. 2013, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Michel, L.O.; Zhang, Q. CmeABC Functions as a Multidrug Efflux System in Campylobacter jejuni. Antimicrob. Agents Chemother. 2002, 46, 2124–2131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiba, M.; Lin, J.; Barton, Y.-W.; Zhang, Q. Interaction of CmeABC and CmeDEF in Conferring Antimicrobial Resistance and Maintaining Cell Viability in Campylobacter jejuni. J. Antimicrob. Chemother. 2005, 57, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Pumbwe, L.; Piddock, L.J.V. Identification and Molecular Characterisation of CmeB, a Campylobacter jejuni Multidrug Efflux Pump. FEMS Microbiol. Lett. 2002, 206, 185–189. [Google Scholar] [CrossRef] [Green Version]
- Luo, N.; Pereira, S.; Sahin, O.; Lin, J.; Huang, S.; Michel, L.; Zhang, Q. Enhanced In Vivo Fitness of Fluoroquinolone-Resistant Campylobacter jejuni in the Absence of Antibiotic Selection Pressure. Proc. Natl. Acad. Sci. USA 2005, 102, 541–546. [Google Scholar] [CrossRef] [Green Version]
- Opperman, T.J.; Kwasny, S.M.; Kim, H.-S.; Nguyen, S.T.; Houseweart, C.; D’Souza, S.V.; Walker, G.C.; Peet, N.P.; Nikaido, H.; Bowlin, T.L. Characterization of A Novel Pyranopyridine Inhibitor of the AcrAB Efflux Pump of Escherichia coli. Antimicrob. Agents Chemother. 2013, 58, 722–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, V.B.; Venkataramaiah, M.; Mondal, A.; Rajamohan, G. Functional Characterization of AbeD, an RND-Type Membrane Transporter in Antimicrobial Resistance in Acinetobacter baumannii. PLoS ONE 2015, 10, e0141314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, V.B.; Vaidyanathan, V.; Rajamohan, G. AbuO, a TolC-Like Outer Membrane Protein of Acinetobacter baumannii, Is Involved in Antimicrobial and Oxidative Stress Resistance. Antimicrob. Agents Chemother. 2014, 59, 1236–1245. [Google Scholar] [CrossRef] [Green Version]
- Blanco, P.; Corona, F.; Martínez, J.L. Involvement of the RND Efflux Pump Transporter SmeH in the Acquisition of Resistance to Ceftazidime in Stenotrophomonas maltophilia. Sci. Rep. 2019, 9, 4917. [Google Scholar] [CrossRef]
- Morgan, J. Pairwise Competition and the Replicator Equation. Bull. Math. Biol. 2003, 65, 1163–1172. [Google Scholar] [CrossRef] [Green Version]
- Jeon, B.; Wang, Y.; Hao, H.; Barton, Y.-W.; Zhang, Q. Contribution of CmeG to Antibiotic and Oxidative Stress Resistance in Campylobacter jejuni. J. Antimicrob. Chemother. 2010, 66, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Lee, B.; Yeom, J.-H.; Hwang, S.; Kang, I.; Cho, J.-C.; Ha, N.-C.; Bae, J.; Lee, K.; Kim, Y.-H. MdsABC-Mediated Pathway for Pathogenicity in Salmonella enterica Serovar Typhimurium. Infect. Immun. 2015, 83, 4266–4276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, E.; Raftery, M.; Goodchild, A.; Mendz, G.L. Campylobacter jejuni Response to Ox-Bile Stress. FEMS Immunol. Med. Microbiol. 2007, 49, 165–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urdaneta, V.; Casadesús, J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef]
- Gunn, J.S. Mechanisms of Bacterial Resistance and Response to Bile. Microbes Infect. 2000, 2, 907–913. [Google Scholar] [CrossRef]
- Lin, J.; Cagliero, C.; Guo, B.; Barton, Y.-W.; Maurel, M.-C.; Payot, S.; Zhang, Q. Bile Salts Modulate Expression of the CmeABC Multidrug Efflux Pump in Campylobacter jejuni. J. Bacteriol. 2005, 187, 7417–7424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pumbwe, L.; Skilbeck, C.A.; Nakano, V.; Avila-Campos, M.J.; Piazza, R.M.; Wexler, H.M. Bile Salts Enhance Bacterial Co-Aggregation, Bacterial-Intestinal Epithelial Cell Adhesion, Biofilm Formation and Antimicrobial Resistance of Bacteroides fragilis. Microb. Pathog. 2007, 43, 78–87. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Houry, A.; Gohar, M.; Deschamps, J.; Tischenko, E.; Aymerich, S.; Gruss, A.; Briandet, R. Bacterial Swimmers that Infiltrate and Take Over the Biofilm Matrix. Proc. Natl. Acad. Sci. USA 2012, 109, 13088–13093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webber, M.A.; Bailey, A.M.; Blair, J.M.A.; Morgan, E.; Stevens, M.P.; Hinton, J.C.D.; Ivens, A.; Wain, J.; Piddock, L.J.V. The Global Consequence of Disruption of the AcrAB-TolC Efflux Pump in Salmonella enterica Includes Reduced Expression of SPI-1 and Other Attributes Required to Infect the Host. J. Bacteriol. 2009, 191, 4276–4285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kvist, M.; Hancock, V.; Klemm, P. Inactivation of Efflux Pumps Abolishes Bacterial Biofilm Formation. Appl. Environ. Microbiol. 2008, 74, 7376–7382. [Google Scholar] [CrossRef] [Green Version]
- Baugh, S.; Phillips, C.R.; Ekanayaka, A.S.; Piddock, L.; Webber, M.A. Inhibition of Multidrug Efflux as A Strategy to Prevent Biofilm Formation. J. Antimicrob. Chemother. 2014, 69, 673–681. [Google Scholar] [CrossRef] [Green Version]
- Yoon, E.-J.; Chabane, Y.N.; Goussard, S.; Snesrud, E.; Courvalin, P.; Dé, E.; Grillot-Courvalin, C. Contribution of Resistance-Nodulation-Cell Division Efflux Systems to Antibiotic Resistance and Biofilm Formation in Acinetobacter baumannii. mBio 2015, 6, e00309-15. [Google Scholar] [CrossRef] [Green Version]
- Baugh, S.; Ekanayaka, A.S.; Piddock, L.; Webber, M.A. Loss of or Inhibition of all Multidrug Resistance Efflux Pumps of Salmonella enterica Serovar Typhimurium Results in Impaired Ability to Form a Biofilm. J. Antimicrob. Chemother. 2012, 67, 2409–2417. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-T.; Huang, Y.-W.; Chen, S.-J.; Chang, C.-W.; Yang, T.-C. The SmeYZ Efflux Pump of Stenotrophomonas maltophilia Contributes to Drug Resistance, Virulence-Related Characteristics, and Virulence in Mice. Antimicrob. Agents Chemother. 2015, 59, 4067–4073. [Google Scholar] [CrossRef] [Green Version]
- Poole, K. Bacterial Multidrug Efflux Pumps Serve Other Functions. Microbe 2008, 3, 179–185. [Google Scholar] [CrossRef]
- Pizarro-Cerdá, J.; Cossart, P. Bacterial Adhesion and Entry into Host Cells. Cell 2006, 124, 715–727. [Google Scholar] [CrossRef] [Green Version]
- Spaniol, V.; Bernhard, S.; Aebi, C. Moraxella catarrhalis AcrAB-OprM Efflux Pump Contributes to Antimicrobial Resistance and Is Enhanced during Cold Shock Response. Antimicrob. Agents Chemother. 2015, 59, 1886–1894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bina, X.R.; Provenzano, D.; Nguyen, N.; Bina, J.E. Vibrio cholerae RND Family Efflux Systems Are Required for Antimicrobial Resistance, Optimal Virulence Factor Production, and Colonization of the Infant Mouse Small Intestine. Infect. Immun. 2008, 76, 3595–3605. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, S.; Fraqueza, M.J.; Queiroz, J.; Domingues, F.C.; Oleastro, M. Genetic Diversity, Antibiotic Resistance and Biofilm-Forming Ability of Arcobacter butzleri Isolated from Poultry and Environment from a Portuguese Slaughterhouse. Int. J. Food Microbiol. 2013, 162, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Bonifácio, M.; Mateus, C.; Alves, A.; Maldonado, E.; Duarte, A.; Domingues, F.; Oleastro, M.; Ferreira, S. Natural Transformation as A Mechanism of Horizontal Gene Transfer in Aliarcobacter butzleri. Pathogens. submitted.
- Ferreira, S.; Correia, D.R.; Oleastro, M.; Domingues, F.C. Arcobacter butzleri Ciprofloxacin Resistance: Point Mutations in DNA Gyrase A and Role on Fitness Cost. Microb. Drug Resist. 2018, 24, 915–922. [Google Scholar] [CrossRef]
- Reeser, R.J.; Medler, R.T.; Billington, S.J.; Jost, B.H.; Joens, L.A. Characterization of Campylobacter jejuni Biofilms under Defined Growth Conditions. Appl. Environ. Microbiol. 2007, 73, 1908–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Shaughnessy, C.M.; Cunningham, A.F.; MacLennan, C.A. The Stability of Complement-Mediated Bactericidal Activity in Human Serum against Salmonella. PLoS ONE 2012, 7, e49147. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Antimicrobial | MIC (μg/mL) [Fold Reduction] | |||
|---|---|---|---|---|
| Ab_2811 | Ab_2811∆areB | Ab_2811∆areE | Ab_2811∆areG | |
| β-Lactams | ||||
| Ampicillin | 128 | 128 [ND] | 128 [ND] | 64 [2] |
| Cephalexin | 256 | 8 [32] | 64 [4] | 0.0625 [4096] |
| Cefotaxime | 64 | 2 [32] | 32 [2] | 0.0625 [1024] |
| Amoxycillin | 64 | 64 [ND] | 64 [ND] | 16 [4] |
| Macrolides | ||||
| Erythromycin | 16 | 2 [8] | 8 [2] | 16 [ND] |
| Clarithromycin | 16 | 1 [16] | 8 [2] | 16 [ND] |
| Aminoglycosides | ||||
| Gentamycin | 1 | 1 [ND] | 1 [ND] | 1 [ND] |
| Kanamycin * | 4 | >512 | >512 | >512 |
| Streptomycin | 8 | 0.5 [16] | 4 [2] | 2 [4] |
| Quinolones | ||||
| Nalidixic acid | >256 | >256 [ND] | >256 [ND] | >256 [ND] |
| Ciprofloxacin | 32 | 8 [4] | 32 [ND] | 16 [4] |
| Levofloxacin | 64 | 32 [2] | 64 [ND] | 16 [4] |
| Moxifloxacin | 32 | 8 [4] | 32 [ND] | 32 [ND] |
| Tetracyclines | ||||
| Tetracyclines | 8 | 8 [ND] | 8 [ND] | 0.5 [16] |
| Doxycycline | 8 | 4 [2] | 4 [2] | 1 [8] |
| Phenicol | ||||
| Chloramphenicol | 32 | 32 [ND] | 32 [ND] | 8 [4] |
| Bile salts | ||||
| Sodium cholate | 8000 | 8000 [ND] | 4000 [2] | 4000 [2] |
| Sodium deoxycholate | 32,000 | 32,000 [ND] | 32,000 [ND] | 32,000 [ND] |
| Bile salts mix | 50,000 | 50,000 [ND] | 50,000 [ND] | 6250 [8] |
| Disinfectants | ||||
| Chlorhexidine | 2 | 2 [ND] | 2 [ND] | 2 [ND] |
| Benzalkonium chloride | 32 | 32 [ND] | 32 [ND] | 16 [2] |
| Germicide | ||||
| Acriflavine | 32 | 16 [2] | 32 [ND] | 16 [2] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mateus, C.; Nunes, A.R.; Oleastro, M.; Domingues, F.; Ferreira, S. RND Efflux Systems Contribute to Resistance and Virulence of Aliarcobacter butzleri. Antibiotics 2021, 10, 823. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10070823
Mateus C, Nunes AR, Oleastro M, Domingues F, Ferreira S. RND Efflux Systems Contribute to Resistance and Virulence of Aliarcobacter butzleri. Antibiotics. 2021; 10(7):823. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10070823
Chicago/Turabian StyleMateus, Cristiana, Ana Rita Nunes, Mónica Oleastro, Fernanda Domingues, and Susana Ferreira. 2021. "RND Efflux Systems Contribute to Resistance and Virulence of Aliarcobacter butzleri" Antibiotics 10, no. 7: 823. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10070823







